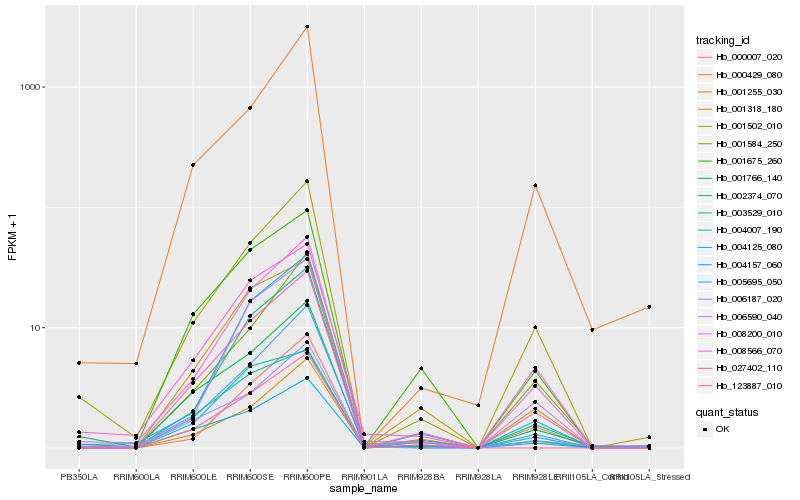
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027402_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 2 |
Hb_001766_140 |
0.0779998359 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 3 |
Hb_001675_260 |
0.0793508817 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 4 |
Hb_008200_010 |
0.0808188125 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001502_010 |
0.0897634103 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 6 |
Hb_001255_030 |
0.0931746745 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840, partial [Sesamum indicum] |
| 7 |
Hb_005695_050 |
0.0967166573 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 8 |
Hb_004157_060 |
0.0983975971 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Jatropha curcas] |
| 9 |
Hb_001318_180 |
0.102386945 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 1 [Jatropha curcas] |
| 10 |
Hb_123887_010 |
0.107096366 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 11 |
Hb_004125_080 |
0.1093594897 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 12 |
Hb_000007_020 |
0.1103210579 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 13 |
Hb_008566_070 |
0.1104885233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006187_020 |
0.1110909949 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Populus euphratica] |
| 15 |
Hb_006590_040 |
0.1142996138 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 16 |
Hb_003529_010 |
0.1143690203 |
- |
- |
PREDICTED: disease resistance protein RPS6-like [Jatropha curcas] |
| 17 |
Hb_002374_070 |
0.1144723255 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_004007_190 |
0.1227557951 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 19 |
Hb_001584_250 |
0.1228029673 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 20 |
Hb_000429_080 |
0.1261347402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |