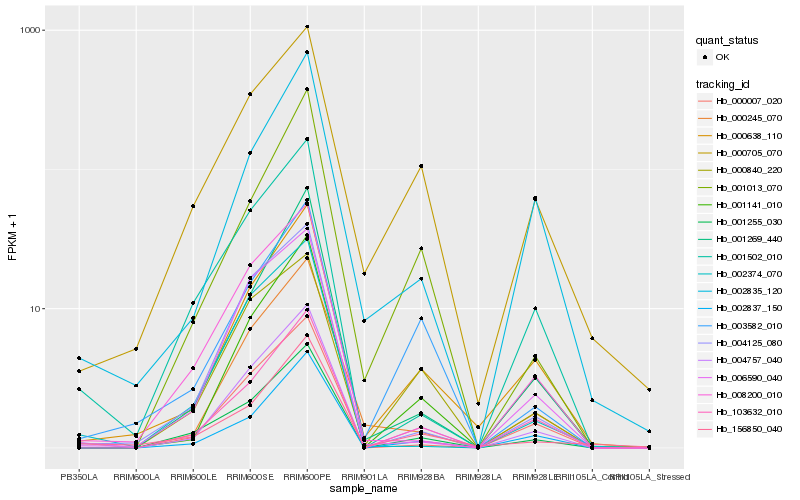
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004757_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_103632_010 |
0.098616724 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 3 |
Hb_000638_110 |
0.1025431988 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0007s04250g [Populus trichocarpa] |
| 4 |
Hb_001141_010 |
0.1041754552 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 5 |
Hb_001255_030 |
0.1059765778 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840, partial [Sesamum indicum] |
| 6 |
Hb_000245_070 |
0.1122946404 |
- |
- |
PREDICTED: uncharacterized protein LOC105141276 [Populus euphratica] |
| 7 |
Hb_002837_150 |
0.1131784992 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 8 |
Hb_000007_020 |
0.1186471487 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 9 |
Hb_004125_080 |
0.1206950725 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 10 |
Hb_000840_220 |
0.1311316671 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 11 |
Hb_001269_440 |
0.1319755333 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 12 |
Hb_001502_010 |
0.1322588996 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 13 |
Hb_002374_070 |
0.1378035176 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_006590_040 |
0.1383668615 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 15 |
Hb_003582_010 |
0.1388651001 |
- |
- |
hypothetical protein EUGRSUZ_B026451, partial [Eucalyptus grandis] |
| 16 |
Hb_008200_010 |
0.1392346373 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002835_120 |
0.1417549672 |
- |
- |
PREDICTED: uncharacterized protein LOC105640304 [Jatropha curcas] |
| 18 |
Hb_156850_040 |
0.1424511827 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 19 |
Hb_000705_070 |
0.1427377453 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001013_070 |
0.143352579 |
- |
- |
- |