| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
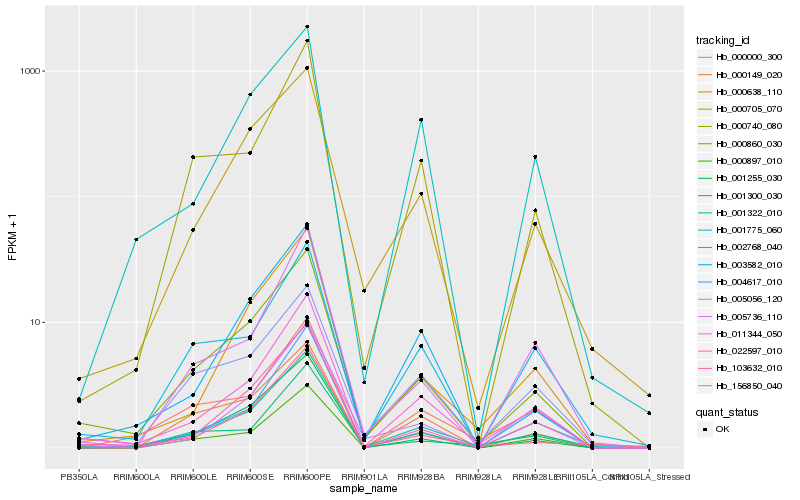
Hb_001322_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 2 |
Hb_022597_010 |
0.0673927362 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 3 |
Hb_000860_030 |
0.0855249506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000705_070 |
0.1063136749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_156850_040 |
0.1094876426 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 6 |
Hb_000638_110 |
0.1111260006 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0007s04250g [Populus trichocarpa] |
| 7 |
Hb_000740_080 |
0.1246321029 |
- |
- |
- |
| 8 |
Hb_003582_010 |
0.1262662376 |
- |
- |
hypothetical protein EUGRSUZ_B026451, partial [Eucalyptus grandis] |
| 9 |
Hb_005056_120 |
0.1277510085 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_000000_300 |
0.1301750042 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 11 |
Hb_001775_060 |
0.1303878939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000897_010 |
0.1309290255 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 13 |
Hb_002768_040 |
0.1323757298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_103632_010 |
0.1341296993 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 15 |
Hb_004617_010 |
0.1346784539 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X3 [Citrus sinensis] |
| 16 |
Hb_011344_050 |
0.1407647907 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 17 |
Hb_001255_030 |
0.1422246714 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840, partial [Sesamum indicum] |
| 18 |
Hb_000149_020 |
0.1438687776 |
- |
- |
Proline-rich receptor-like protein kinase PERK14 [Glycine soja] |
| 19 |
Hb_001300_030 |
0.143969155 |
- |
- |
hypothetical protein JCGZ_13871 [Jatropha curcas] |
| 20 |
Hb_005736_110 |
0.1496994046 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |