| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
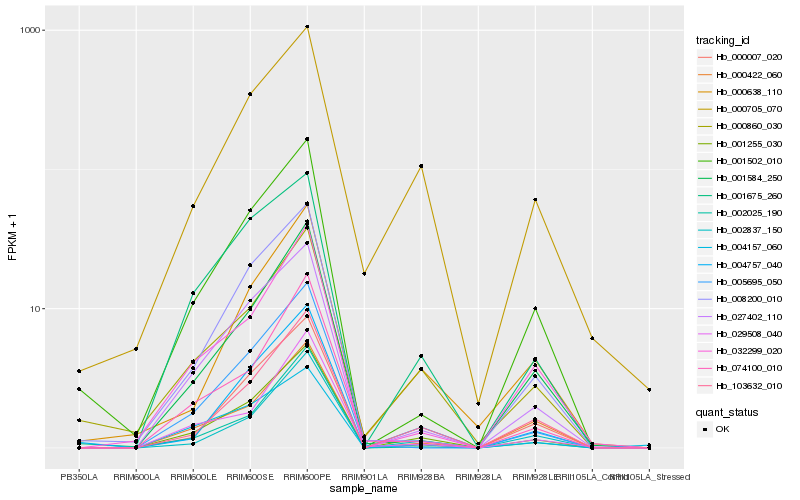
Hb_001255_030 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840, partial [Sesamum indicum] |
| 2 |
Hb_103632_010 |
0.0900224724 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 3 |
Hb_027402_110 |
0.0931746745 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 4 |
Hb_004757_040 |
0.1059765778 |
- |
- |
- |
| 5 |
Hb_002837_150 |
0.1138124695 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 6 |
Hb_001675_260 |
0.1191935081 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 7 |
Hb_000860_030 |
0.1195926954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001502_010 |
0.121604109 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 9 |
Hb_001584_250 |
0.1228203202 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 10 |
Hb_000007_020 |
0.1245866515 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 11 |
Hb_074100_010 |
0.1247389998 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 12 |
Hb_032299_020 |
0.1248537857 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 13 |
Hb_002025_190 |
0.1268957074 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 14 |
Hb_005695_050 |
0.1294045209 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 15 |
Hb_008200_010 |
0.1294778789 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_029508_040 |
0.1303812311 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 17 |
Hb_000422_060 |
0.1304212011 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 18 |
Hb_000638_110 |
0.1305841272 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0007s04250g [Populus trichocarpa] |
| 19 |
Hb_000705_070 |
0.1351277935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004157_060 |
0.1353221927 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Jatropha curcas] |