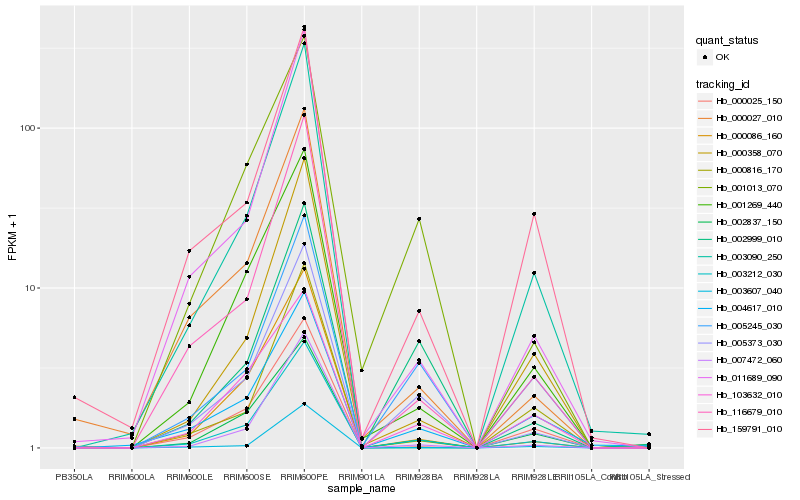
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003212_030 |
0.0 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 2 |
Hb_005373_030 |
0.0860600737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002837_150 |
0.0964283103 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 4 |
Hb_000086_160 |
0.0982184184 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 5 |
Hb_001269_440 |
0.1005086211 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 6 |
Hb_004617_010 |
0.108874136 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X3 [Citrus sinensis] |
| 7 |
Hb_001013_070 |
0.1142178828 |
- |
- |
- |
| 8 |
Hb_116679_010 |
0.1144837147 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 9 |
Hb_000358_070 |
0.1155865586 |
- |
- |
hypothetical protein POPTR_0003s01410g [Populus trichocarpa] |
| 10 |
Hb_007472_060 |
0.1160107353 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000025_150 |
0.1199113423 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 12 |
Hb_159791_010 |
0.1261091185 |
- |
- |
PREDICTED: cytochrome P450 84A1 [Jatropha curcas] |
| 13 |
Hb_000027_010 |
0.1267129347 |
- |
- |
- |
| 14 |
Hb_011689_090 |
0.1285115508 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 15 |
Hb_005245_030 |
0.1293705897 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g07570 [Jatropha curcas] |
| 16 |
Hb_103632_010 |
0.1315258967 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 17 |
Hb_003607_040 |
0.1334012697 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 18 |
Hb_002999_010 |
0.13362114 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 19 |
Hb_003090_250 |
0.1341438077 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000816_170 |
0.1342354593 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |