| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
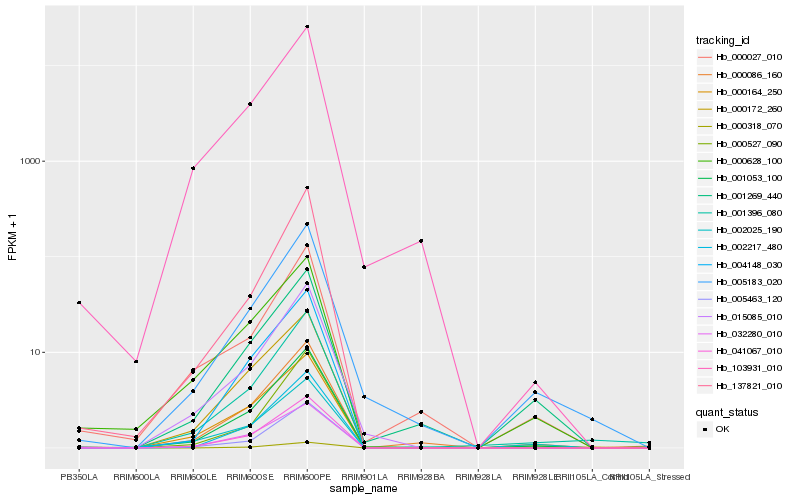
Hb_001053_100 |
0.0 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 2 |
Hb_000086_160 |
0.0646472615 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 3 |
Hb_103931_010 |
0.074472512 |
- |
- |
- |
| 4 |
Hb_041067_010 |
0.0756116785 |
- |
- |
hypothetical protein JCGZ_00780 [Jatropha curcas] |
| 5 |
Hb_015085_010 |
0.0819571792 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 6 |
Hb_001396_080 |
0.0829195876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005183_020 |
0.0837615746 |
- |
- |
hypothetical protein JCGZ_04285 [Jatropha curcas] |
| 8 |
Hb_032280_010 |
0.0912348198 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004148_030 |
0.0928411087 |
- |
- |
PREDICTED: protein RAFTIN 1B-like [Gossypium raimondii] |
| 10 |
Hb_002217_480 |
0.0942617071 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 11 |
Hb_000172_260 |
0.0976154198 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 12 |
Hb_002025_190 |
0.0990697652 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 13 |
Hb_000027_010 |
0.1009033993 |
- |
- |
- |
| 14 |
Hb_001269_440 |
0.1027643372 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 15 |
Hb_000164_250 |
0.1035727206 |
- |
- |
hypothetical protein CISIN_1g040645mg [Citrus sinensis] |
| 16 |
Hb_005463_120 |
0.1037519522 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 17 |
Hb_000527_090 |
0.1049845673 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 18 |
Hb_000628_100 |
0.1058838034 |
- |
- |
hypothetical protein MTR_1g019660 [Medicago truncatula] |
| 19 |
Hb_137821_010 |
0.1083146704 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |
| 20 |
Hb_000318_070 |
0.11068895 |
- |
- |
PREDICTED: peroxidase 10-like [Jatropha curcas] |