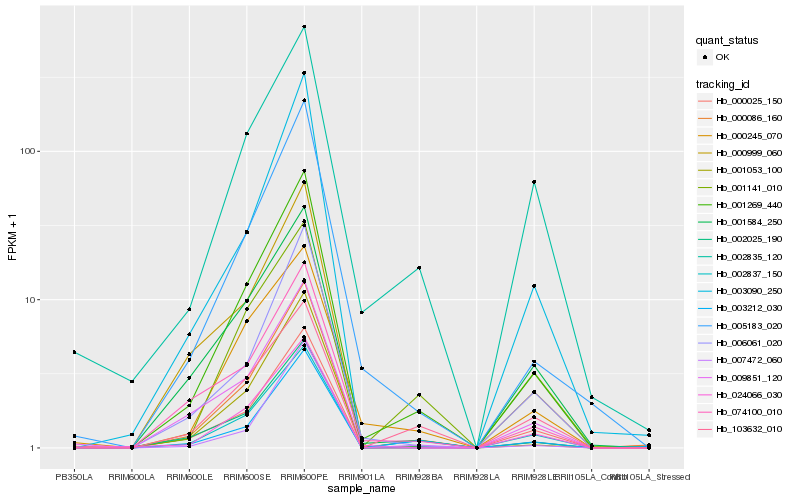
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_440 |
0.0 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 2 |
Hb_002837_150 |
0.0794770139 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 3 |
Hb_000086_160 |
0.0880146111 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 4 |
Hb_002025_190 |
0.0889254324 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 5 |
Hb_005183_020 |
0.0932764411 |
- |
- |
hypothetical protein JCGZ_04285 [Jatropha curcas] |
| 6 |
Hb_000025_150 |
0.1002590768 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 7 |
Hb_003212_030 |
0.1005086211 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 8 |
Hb_001053_100 |
0.1027643372 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 9 |
Hb_024066_030 |
0.1034129583 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 10 |
Hb_006061_020 |
0.1042543815 |
transcription factor |
TF Family: bZIP |
bZIP family protein [Populus trichocarpa] |
| 11 |
Hb_001584_250 |
0.109438824 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 12 |
Hb_074100_010 |
0.1120058375 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 13 |
Hb_001141_010 |
0.1155976659 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 14 |
Hb_003090_250 |
0.1167743429 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000999_060 |
0.117421239 |
- |
- |
hypothetical protein POPTR_0018s11350g [Populus trichocarpa] |
| 16 |
Hb_000245_070 |
0.1180878927 |
- |
- |
PREDICTED: uncharacterized protein LOC105141276 [Populus euphratica] |
| 17 |
Hb_103632_010 |
0.118313156 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 18 |
Hb_007472_060 |
0.1211036215 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_009851_120 |
0.1214870396 |
- |
- |
PREDICTED: RING-H2 zinc finger protein RHA4a [Jatropha curcas] |
| 20 |
Hb_002835_120 |
0.1214886729 |
- |
- |
PREDICTED: uncharacterized protein LOC105640304 [Jatropha curcas] |