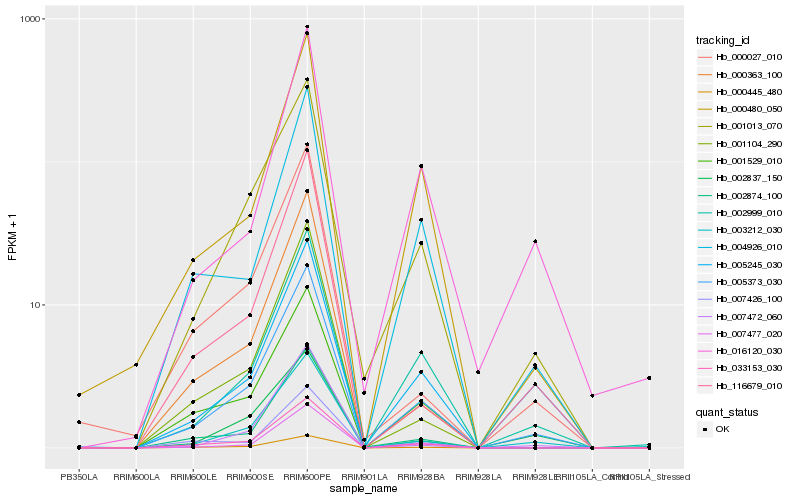
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005373_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001013_070 |
0.0780930797 |
- |
- |
- |
| 3 |
Hb_002999_010 |
0.0799125713 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 4 |
Hb_003212_030 |
0.0860600737 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_033153_030 |
0.0875543315 |
- |
- |
PREDICTED: uncharacterized protein LOC105650145 [Jatropha curcas] |
| 6 |
Hb_000445_480 |
0.1006346341 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_007477_020 |
0.1082267298 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA5 [Jatropha curcas] |
| 8 |
Hb_001529_010 |
0.1099810538 |
- |
- |
- |
| 9 |
Hb_005245_030 |
0.1126532236 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g07570 [Jatropha curcas] |
| 10 |
Hb_000480_050 |
0.1150317526 |
- |
- |
hypothetical protein PHAVU_004G154200g [Phaseolus vulgaris] |
| 11 |
Hb_002874_100 |
0.1159999071 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_004926_010 |
0.1192402525 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 13 |
Hb_000363_100 |
0.1281351814 |
- |
- |
- |
| 14 |
Hb_001104_290 |
0.1294459279 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 15 |
Hb_116679_010 |
0.1294537346 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 16 |
Hb_007426_100 |
0.1307443782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002837_150 |
0.1324791614 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 18 |
Hb_000027_010 |
0.1336447858 |
- |
- |
- |
| 19 |
Hb_016120_030 |
0.1336785666 |
- |
- |
hypothetical protein EUGRSUZ_D00080 [Eucalyptus grandis] |
| 20 |
Hb_007472_060 |
0.1338045535 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |