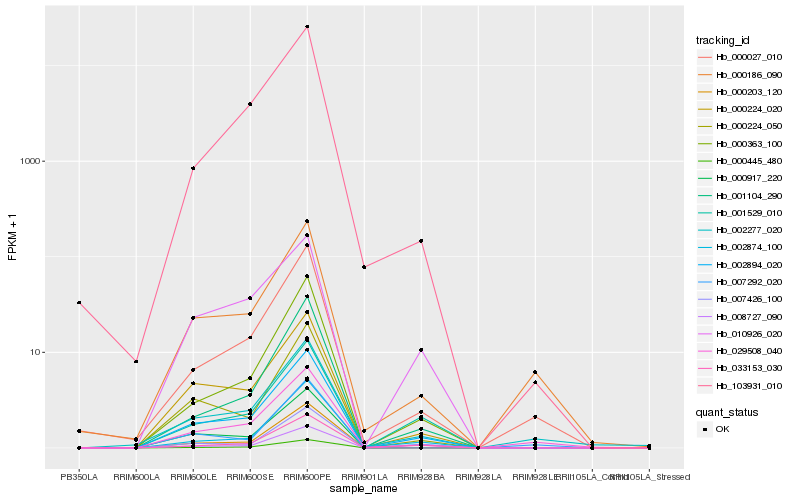
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002894_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 2 |
Hb_000445_480 |
0.0597349099 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_000224_020 |
0.076006141 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 4 |
Hb_033153_030 |
0.0872212866 |
- |
- |
PREDICTED: uncharacterized protein LOC105650145 [Jatropha curcas] |
| 5 |
Hb_007426_100 |
0.0900381847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002874_100 |
0.0975948882 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_000363_100 |
0.100780924 |
- |
- |
- |
| 8 |
Hb_000224_050 |
0.1013361117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001529_010 |
0.1017794281 |
- |
- |
- |
| 10 |
Hb_001104_290 |
0.1061073635 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 11 |
Hb_010926_020 |
0.1076994605 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Camelina sativa] |
| 12 |
Hb_000027_010 |
0.1141465509 |
- |
- |
- |
| 13 |
Hb_000186_090 |
0.119603436 |
- |
- |
ACC oxidase 1 [Hevea brasiliensis] |
| 14 |
Hb_008727_090 |
0.1197033174 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 15 |
Hb_000203_120 |
0.1205853115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000917_220 |
0.1218050276 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 17 |
Hb_002277_020 |
0.1230435929 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |
| 18 |
Hb_007292_020 |
0.1246041784 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 19 |
Hb_029508_040 |
0.1246592124 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 20 |
Hb_103931_010 |
0.1267853367 |
- |
- |
- |