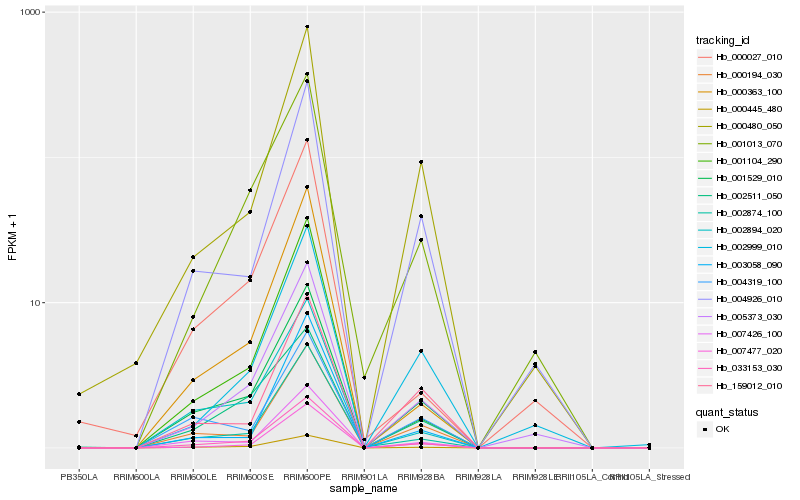
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033153_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650145 [Jatropha curcas] |
| 2 |
Hb_000445_480 |
0.0320533063 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_001529_010 |
0.0565078816 |
- |
- |
- |
| 4 |
Hb_002874_100 |
0.0642399564 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_007426_100 |
0.0692262299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002894_020 |
0.0872212866 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 7 |
Hb_005373_030 |
0.0875543315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000363_100 |
0.0887715042 |
- |
- |
- |
| 9 |
Hb_001104_290 |
0.0922741058 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 10 |
Hb_000194_030 |
0.0951186019 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 11 |
Hb_007477_020 |
0.0962073582 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA5 [Jatropha curcas] |
| 12 |
Hb_001013_070 |
0.1165376633 |
- |
- |
- |
| 13 |
Hb_003058_090 |
0.1176919813 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 14 |
Hb_000480_050 |
0.1200372437 |
- |
- |
hypothetical protein PHAVU_004G154200g [Phaseolus vulgaris] |
| 15 |
Hb_004926_010 |
0.1230028437 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 16 |
Hb_002999_010 |
0.1297416936 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 17 |
Hb_002511_050 |
0.1299633615 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000027_010 |
0.1308130283 |
- |
- |
- |
| 19 |
Hb_159012_010 |
0.1327181865 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 20 |
Hb_004319_100 |
0.1332410493 |
- |
- |
PREDICTED: gibberellin-regulated protein 14 isoform X3 [Jatropha curcas] |