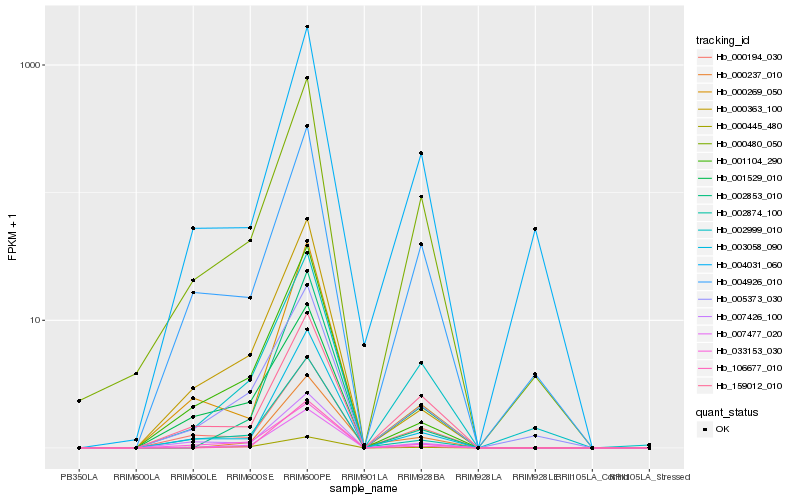
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007477_020 |
0.0 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA5 [Jatropha curcas] |
| 2 |
Hb_003058_090 |
0.0760513957 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 3 |
Hb_000480_050 |
0.09025175 |
- |
- |
hypothetical protein PHAVU_004G154200g [Phaseolus vulgaris] |
| 4 |
Hb_033153_030 |
0.0962073582 |
- |
- |
PREDICTED: uncharacterized protein LOC105650145 [Jatropha curcas] |
| 5 |
Hb_002999_010 |
0.1007399619 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 6 |
Hb_002874_100 |
0.1038369663 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_005373_030 |
0.1082267298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000194_030 |
0.112316665 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 9 |
Hb_004926_010 |
0.1147518232 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 10 |
Hb_159012_010 |
0.1187044681 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 11 |
Hb_007426_100 |
0.1189450661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000269_050 |
0.1199861216 |
- |
- |
PREDICTED: beta-glucosidase 12-like [Gossypium raimondii] |
| 13 |
Hb_000237_010 |
0.1241550464 |
transcription factor |
TF Family: SRS |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001104_290 |
0.1242411873 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 15 |
Hb_000363_100 |
0.1249491136 |
- |
- |
- |
| 16 |
Hb_000445_480 |
0.1260559804 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_001529_010 |
0.1275138323 |
- |
- |
- |
| 18 |
Hb_002853_010 |
0.1324759467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004031_060 |
0.1325317666 |
- |
- |
hypothetical protein B456_007G3219002, partial [Gossypium raimondii] |
| 20 |
Hb_106677_010 |
0.1351807078 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |