| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
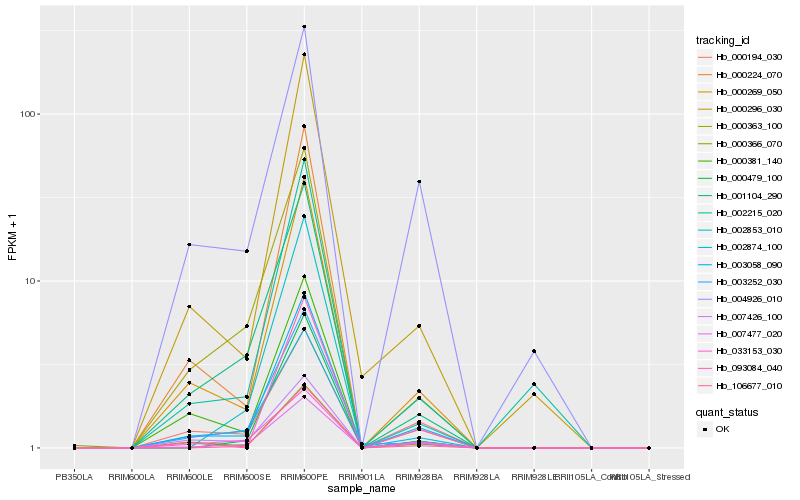
Hb_003058_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 2 |
Hb_000269_050 |
0.0459762971 |
- |
- |
PREDICTED: beta-glucosidase 12-like [Gossypium raimondii] |
| 3 |
Hb_007477_020 |
0.0760513957 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA5 [Jatropha curcas] |
| 4 |
Hb_002874_100 |
0.0929053383 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_007426_100 |
0.1018912144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000296_030 |
0.1023269516 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 7 |
Hb_001104_290 |
0.109600275 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 8 |
Hb_000363_100 |
0.1105572246 |
- |
- |
- |
| 9 |
Hb_093084_040 |
0.1132634507 |
transcription factor |
TF Family: LOB |
hypothetical protein POPTR_0015s09370g [Populus trichocarpa] |
| 10 |
Hb_000381_140 |
0.1163117917 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 11 |
Hb_106677_010 |
0.1163829581 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 12 |
Hb_033153_030 |
0.1176919813 |
- |
- |
PREDICTED: uncharacterized protein LOC105650145 [Jatropha curcas] |
| 13 |
Hb_003252_030 |
0.1185572937 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 14 |
Hb_000194_030 |
0.12024316 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 15 |
Hb_000479_100 |
0.1202480139 |
- |
- |
Alpha-1,4 glucan phosphorylase L-2 isozyme, chloroplastic/amyloplastic [Gossypium arboreum] |
| 16 |
Hb_002853_010 |
0.1231076378 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000224_070 |
0.125289693 |
- |
- |
- |
| 18 |
Hb_000366_070 |
0.1267622114 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.2 [Jatropha curcas] |
| 19 |
Hb_002215_020 |
0.127548754 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 20 |
Hb_004926_010 |
0.1282000127 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |