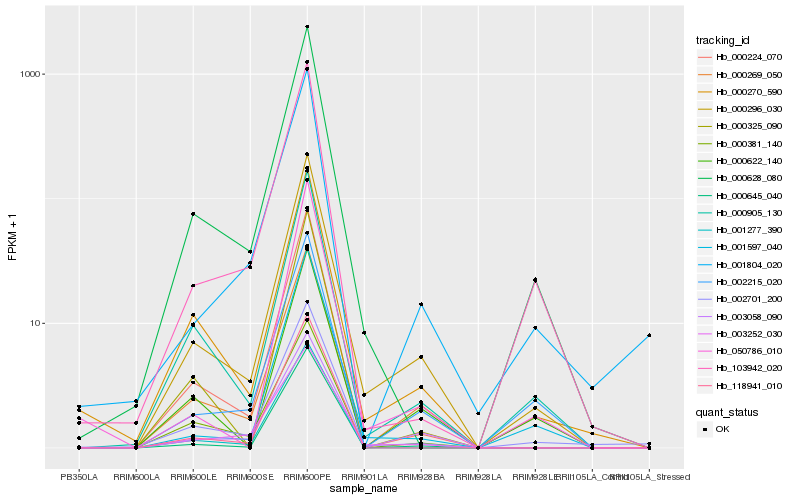
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000296_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 2 |
Hb_000905_130 |
0.0829043498 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100 [Jatropha curcas] |
| 3 |
Hb_000269_050 |
0.0848166061 |
- |
- |
PREDICTED: beta-glucosidase 12-like [Gossypium raimondii] |
| 4 |
Hb_000270_590 |
0.0964897953 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 5 |
Hb_000224_070 |
0.0996843052 |
- |
- |
- |
| 6 |
Hb_001597_040 |
0.1010864979 |
- |
- |
- |
| 7 |
Hb_002701_200 |
0.1018653105 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 8 |
Hb_003058_090 |
0.1023269516 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 9 |
Hb_002215_020 |
0.1040783108 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 10 |
Hb_000628_080 |
0.1043325725 |
- |
- |
hypothetical protein JCGZ_04283 [Jatropha curcas] |
| 11 |
Hb_000645_040 |
0.1054563681 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 12 |
Hb_003252_030 |
0.1076678224 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 13 |
Hb_000622_140 |
0.1140991879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000381_140 |
0.1154665967 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 15 |
Hb_050786_010 |
0.1204238436 |
- |
- |
hypothetical protein MIMGU_mgv1a014824mg [Erythranthe guttata] |
| 16 |
Hb_000325_090 |
0.1209801503 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE5-like [Gossypium raimondii] |
| 17 |
Hb_001277_390 |
0.1222772856 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 18 |
Hb_118941_010 |
0.1244377322 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 19 |
Hb_001804_020 |
0.1246942671 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 20 |
Hb_103942_020 |
0.125316244 |
- |
- |
unknown [Zea mays] |