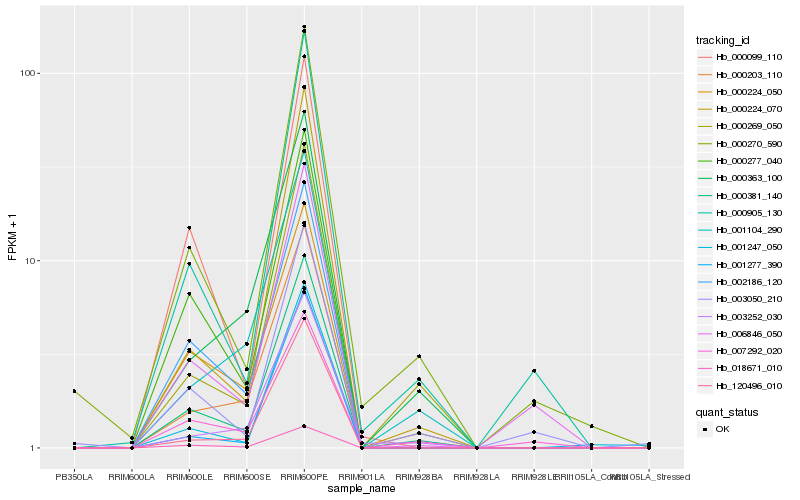
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000381_140 |
0.0 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 2 |
Hb_000269_050 |
0.0787479264 |
- |
- |
PREDICTED: beta-glucosidase 12-like [Gossypium raimondii] |
| 3 |
Hb_000224_070 |
0.0863663289 |
- |
- |
- |
| 4 |
Hb_000224_050 |
0.0908392767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000905_130 |
0.0927816454 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100 [Jatropha curcas] |
| 6 |
Hb_002186_120 |
0.0940743113 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X5 [Populus euphratica] |
| 7 |
Hb_000277_040 |
0.0959621726 |
- |
- |
- |
| 8 |
Hb_018671_010 |
0.0965890229 |
- |
- |
- |
| 9 |
Hb_000270_590 |
0.0984588946 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 10 |
Hb_120496_010 |
0.0990751332 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Cicer arietinum] |
| 11 |
Hb_000203_110 |
0.1027690267 |
- |
- |
- |
| 12 |
Hb_001277_390 |
0.1085636947 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 13 |
Hb_000363_100 |
0.1089602702 |
- |
- |
- |
| 14 |
Hb_001104_290 |
0.1095585847 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 15 |
Hb_003050_210 |
0.1098165045 |
- |
- |
PREDICTED: cytochrome b561 domain-containing protein At4g18260-like [Populus euphratica] |
| 16 |
Hb_000099_110 |
0.110123495 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 17 |
Hb_003252_030 |
0.111566435 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 18 |
Hb_007292_020 |
0.1125571469 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 19 |
Hb_006846_050 |
0.1130767493 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 20 |
Hb_001247_050 |
0.1142137371 |
- |
- |
PREDICTED: WAT1-related protein At1g09380 [Jatropha curcas] |