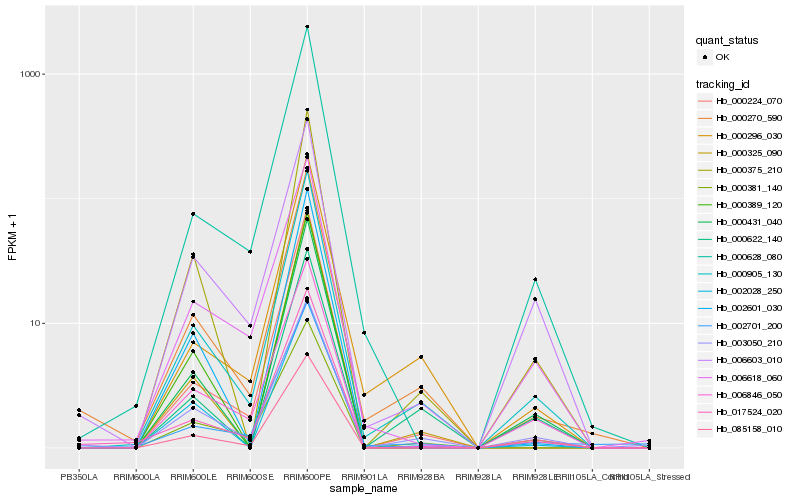
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000905_130 |
0.0 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100 [Jatropha curcas] |
| 2 |
Hb_000270_590 |
0.0707956179 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 3 |
Hb_000375_210 |
0.0739253137 |
- |
- |
- |
| 4 |
Hb_003050_210 |
0.0783184278 |
- |
- |
PREDICTED: cytochrome b561 domain-containing protein At4g18260-like [Populus euphratica] |
| 5 |
Hb_000628_080 |
0.0822794946 |
- |
- |
hypothetical protein JCGZ_04283 [Jatropha curcas] |
| 6 |
Hb_000296_030 |
0.0829043498 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 7 |
Hb_006846_050 |
0.084249403 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 8 |
Hb_000224_070 |
0.0904408938 |
- |
- |
- |
| 9 |
Hb_000431_040 |
0.0905209127 |
- |
- |
PREDICTED: uncharacterized protein LOC105635551 [Jatropha curcas] |
| 10 |
Hb_002701_200 |
0.091314849 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 11 |
Hb_000381_140 |
0.0927816454 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 12 |
Hb_017524_020 |
0.0946484817 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 13 |
Hb_006603_010 |
0.0977518745 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 14 |
Hb_000622_140 |
0.0979032624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002028_250 |
0.0985768449 |
- |
- |
Alpha-aminoadipic semialdehyde synthase, mitochondrial [Crassostrea gigas] |
| 16 |
Hb_002601_030 |
0.0988328042 |
- |
- |
PREDICTED: uncharacterized protein LOC105634908 [Jatropha curcas] |
| 17 |
Hb_085158_010 |
0.0994008199 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 18 |
Hb_000389_120 |
0.1005642794 |
- |
- |
hypothetical protein JCGZ_20066 [Jatropha curcas] |
| 19 |
Hb_000325_090 |
0.1007260877 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE5-like [Gossypium raimondii] |
| 20 |
Hb_006618_060 |
0.1010664197 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |