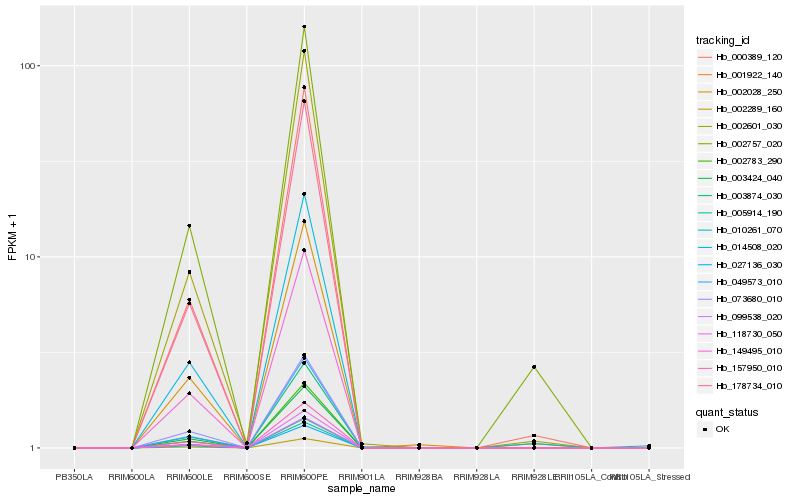
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014508_020 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002028_250 |
0.0435771503 |
- |
- |
Alpha-aminoadipic semialdehyde synthase, mitochondrial [Crassostrea gigas] |
| 3 |
Hb_000389_120 |
0.0473848655 |
- |
- |
hypothetical protein JCGZ_20066 [Jatropha curcas] |
| 4 |
Hb_099538_020 |
0.0486844023 |
- |
- |
hypothetical protein POPTR_0010s12650g [Populus trichocarpa] |
| 5 |
Hb_010261_070 |
0.0486844866 |
- |
- |
hypothetical protein B456_009G094800 [Gossypium raimondii] |
| 6 |
Hb_005914_190 |
0.0489088601 |
- |
- |
PREDICTED: acid phosphatase 1-like [Populus euphratica] |
| 7 |
Hb_002289_160 |
0.0491552289 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 8 |
Hb_027136_030 |
0.0492175338 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_149495_010 |
0.0492935321 |
- |
- |
putative starch phosphorylase, partial [Sorghum bicolor] |
| 10 |
Hb_003874_030 |
0.0501260016 |
- |
- |
hypothetical protein JCGZ_00004 [Jatropha curcas] |
| 11 |
Hb_002601_030 |
0.0511037211 |
- |
- |
PREDICTED: uncharacterized protein LOC105634908 [Jatropha curcas] |
| 12 |
Hb_049573_010 |
0.0515860977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002757_020 |
0.0524501887 |
- |
- |
- |
| 14 |
Hb_001922_140 |
0.0525189149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_178734_010 |
0.0530418251 |
- |
- |
PREDICTED: uncharacterized protein LOC103331175 [Prunus mume] |
| 16 |
Hb_003424_040 |
0.0530747783 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 17 |
Hb_157950_010 |
0.0536030738 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_073680_010 |
0.0536503819 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_002783_290 |
0.0542920474 |
- |
- |
Uncharacterized protein TCM_034216 [Theobroma cacao] |
| 20 |
Hb_118730_050 |
0.054349783 |
- |
- |
RING/U-box superfamily protein isoform 4 [Theobroma cacao] |