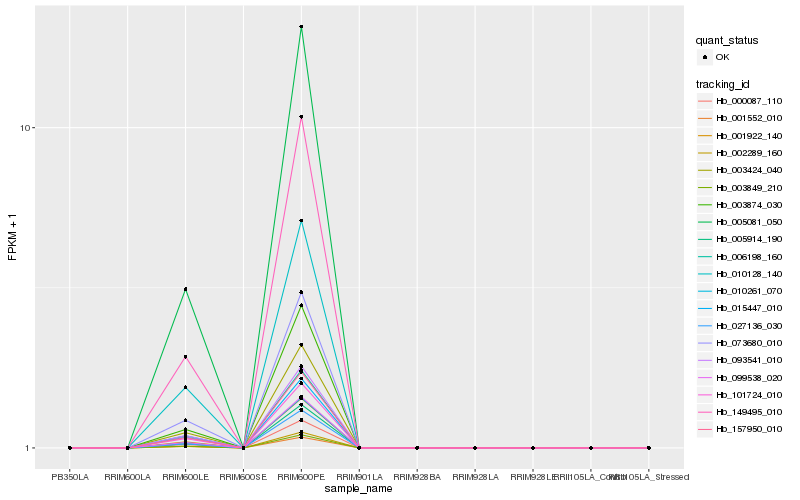
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001922_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003424_040 |
0.0014379372 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 3 |
Hb_157950_010 |
0.0027324149 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_073680_010 |
0.0028453295 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_005081_050 |
0.0046989008 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Prunus mume] |
| 6 |
Hb_000087_110 |
0.0115225508 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_149495_010 |
0.0119409352 |
- |
- |
putative starch phosphorylase, partial [Sorghum bicolor] |
| 8 |
Hb_027136_030 |
0.0124364691 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_002289_160 |
0.0128686116 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 10 |
Hb_093541_010 |
0.0134862768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_101724_010 |
0.0145309937 |
- |
- |
glutamate receptor family protein [Populus trichocarpa] |
| 12 |
Hb_099538_020 |
0.0186120355 |
- |
- |
hypothetical protein POPTR_0010s12650g [Populus trichocarpa] |
| 13 |
Hb_006198_160 |
0.0197952392 |
- |
- |
hypothetical protein POPTR_0002s24290g [Populus trichocarpa] |
| 14 |
Hb_010261_070 |
0.0209044782 |
- |
- |
hypothetical protein B456_009G094800 [Gossypium raimondii] |
| 15 |
Hb_015447_010 |
0.0220041734 |
- |
- |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 16 |
Hb_005914_190 |
0.0245758723 |
- |
- |
PREDICTED: acid phosphatase 1-like [Populus euphratica] |
| 17 |
Hb_003849_210 |
0.0282704377 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0793640 [Ricinus communis] |
| 18 |
Hb_010128_140 |
0.030813235 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 19 |
Hb_003874_030 |
0.0317457317 |
- |
- |
hypothetical protein JCGZ_00004 [Jatropha curcas] |
| 20 |
Hb_001552_010 |
0.0342798415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |