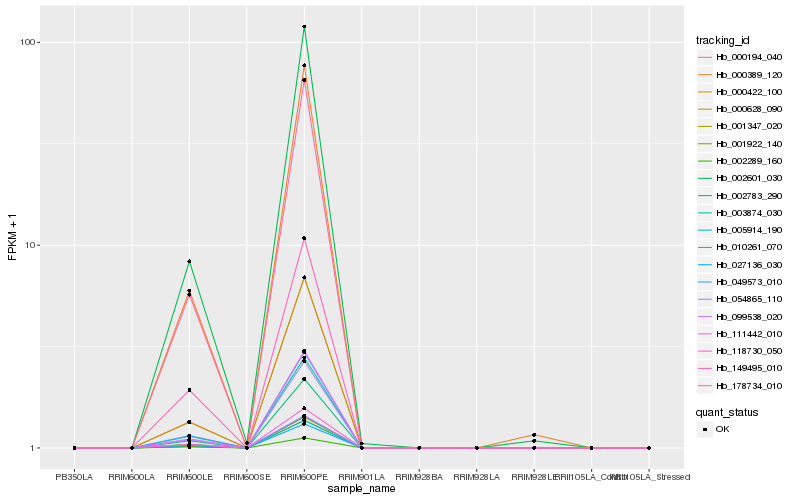
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_178734_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103331175 [Prunus mume] |
| 2 |
Hb_002783_290 |
0.0029793503 |
- |
- |
Uncharacterized protein TCM_034216 [Theobroma cacao] |
| 3 |
Hb_118730_050 |
0.003109678 |
- |
- |
RING/U-box superfamily protein isoform 4 [Theobroma cacao] |
| 4 |
Hb_049573_010 |
0.0039968791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003874_030 |
0.009110521 |
- |
- |
hypothetical protein JCGZ_00004 [Jatropha curcas] |
| 6 |
Hb_005914_190 |
0.0162889963 |
- |
- |
PREDICTED: acid phosphatase 1-like [Populus euphratica] |
| 7 |
Hb_010261_070 |
0.0199614256 |
- |
- |
hypothetical protein B456_009G094800 [Gossypium raimondii] |
| 8 |
Hb_099538_020 |
0.0222533987 |
- |
- |
hypothetical protein POPTR_0010s12650g [Populus trichocarpa] |
| 9 |
Hb_000194_040 |
0.0235240668 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 10 |
Hb_054865_110 |
0.0239914476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000628_090 |
0.0252535421 |
- |
- |
hypothetical protein B456_010G019400 [Gossypium raimondii] |
| 12 |
Hb_002289_160 |
0.0279920056 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 13 |
Hb_027136_030 |
0.0284235803 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_149495_010 |
0.0289184284 |
- |
- |
putative starch phosphorylase, partial [Sorghum bicolor] |
| 15 |
Hb_111442_010 |
0.0307401943 |
desease resistance |
Gene Name: AAA_16 |
LRR and NB-ARC domains-containing disease resistance protein, putative [Theobroma cacao] |
| 16 |
Hb_002601_030 |
0.0327829691 |
- |
- |
PREDICTED: uncharacterized protein LOC105634908 [Jatropha curcas] |
| 17 |
Hb_000422_100 |
0.0335731051 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 18 |
Hb_000389_120 |
0.0338162508 |
- |
- |
hypothetical protein JCGZ_20066 [Jatropha curcas] |
| 19 |
Hb_001347_020 |
0.0348412897 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Populus euphratica] |
| 20 |
Hb_001922_140 |
0.0408320516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |