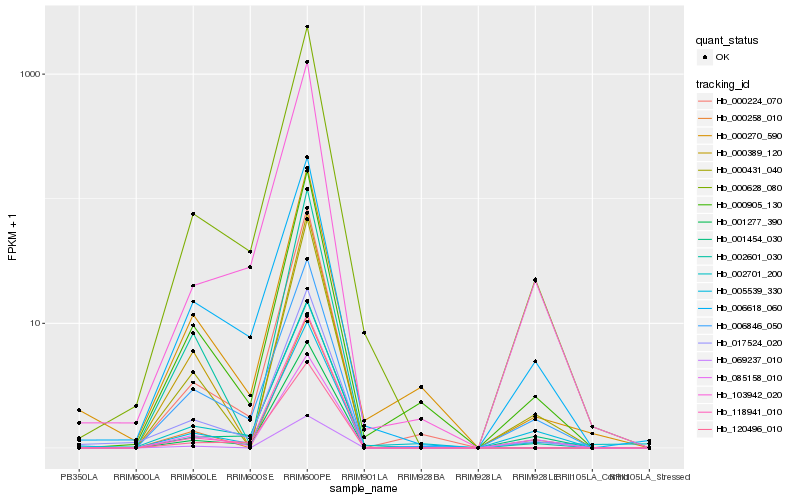
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017524_020 |
0.0 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 2 |
Hb_000628_080 |
0.0740628552 |
- |
- |
hypothetical protein JCGZ_04283 [Jatropha curcas] |
| 3 |
Hb_001277_390 |
0.0939470982 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 4 |
Hb_000905_130 |
0.0946484817 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100 [Jatropha curcas] |
| 5 |
Hb_103942_020 |
0.0978912319 |
- |
- |
unknown [Zea mays] |
| 6 |
Hb_001454_030 |
0.0989599595 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 3 [Jatropha curcas] |
| 7 |
Hb_000224_070 |
0.1001311988 |
- |
- |
- |
| 8 |
Hb_118941_010 |
0.1009991734 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 9 |
Hb_006846_050 |
0.1011790562 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_000431_040 |
0.1014586039 |
- |
- |
PREDICTED: uncharacterized protein LOC105635551 [Jatropha curcas] |
| 11 |
Hb_006618_060 |
0.1047553262 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 12 |
Hb_002701_200 |
0.1061857742 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 13 |
Hb_000270_590 |
0.107316212 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 14 |
Hb_120496_010 |
0.1079922501 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Cicer arietinum] |
| 15 |
Hb_002601_030 |
0.1119331156 |
- |
- |
PREDICTED: uncharacterized protein LOC105634908 [Jatropha curcas] |
| 16 |
Hb_085158_010 |
0.1119681412 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 17 |
Hb_005539_330 |
0.1123630763 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 18 |
Hb_000389_120 |
0.1133572833 |
- |
- |
hypothetical protein JCGZ_20066 [Jatropha curcas] |
| 19 |
Hb_069237_010 |
0.1137789995 |
- |
- |
- |
| 20 |
Hb_000258_010 |
0.1138958898 |
- |
- |
lipid transfer precursor protein [Hevea brasiliensis] |