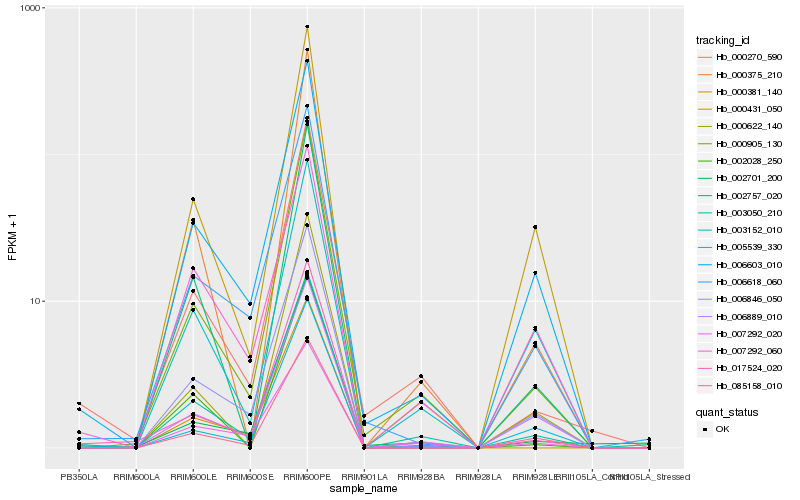
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003050_210 |
0.0 |
- |
- |
PREDICTED: cytochrome b561 domain-containing protein At4g18260-like [Populus euphratica] |
| 2 |
Hb_000905_130 |
0.0783184278 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100 [Jatropha curcas] |
| 3 |
Hb_000270_590 |
0.0823419919 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 4 |
Hb_006603_010 |
0.0889463023 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 5 |
Hb_006846_050 |
0.0977873792 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 6 |
Hb_000375_210 |
0.1030510836 |
- |
- |
- |
| 7 |
Hb_002701_200 |
0.1057175962 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 8 |
Hb_006889_010 |
0.1080625203 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 9 |
Hb_006618_060 |
0.1085217019 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 10 |
Hb_000381_140 |
0.1098165045 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 11 |
Hb_007292_020 |
0.1111801081 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 12 |
Hb_005539_330 |
0.1118888452 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 13 |
Hb_000622_140 |
0.113580174 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_085158_010 |
0.1143626693 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 15 |
Hb_003152_010 |
0.115025826 |
- |
- |
- |
| 16 |
Hb_002028_250 |
0.1159401411 |
- |
- |
Alpha-aminoadipic semialdehyde synthase, mitochondrial [Crassostrea gigas] |
| 17 |
Hb_002757_020 |
0.1204565245 |
- |
- |
- |
| 18 |
Hb_017524_020 |
0.1210856278 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 19 |
Hb_000431_050 |
0.1211511777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007292_060 |
0.1217495575 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |