| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
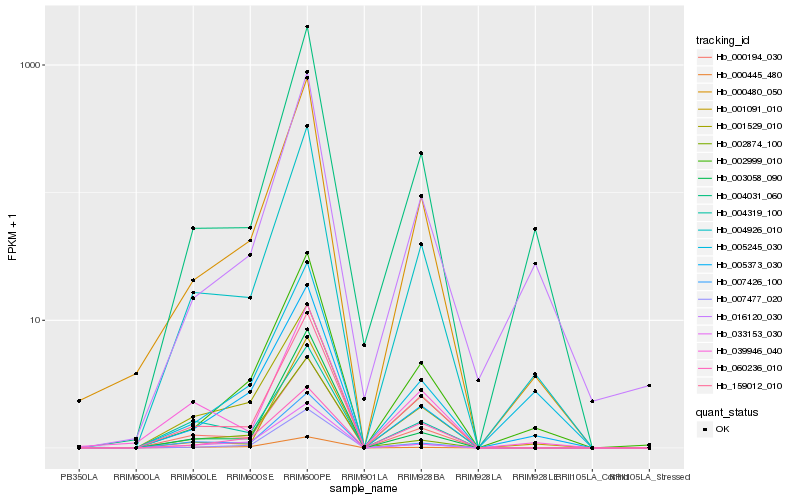
Hb_000480_050 |
0.0 |
- |
- |
hypothetical protein PHAVU_004G154200g [Phaseolus vulgaris] |
| 2 |
Hb_004926_010 |
0.0720286097 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 3 |
Hb_159012_010 |
0.0812644112 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 4 |
Hb_002999_010 |
0.0839095841 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 5 |
Hb_007477_020 |
0.09025175 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA5 [Jatropha curcas] |
| 6 |
Hb_000194_030 |
0.0933986605 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 7 |
Hb_004031_060 |
0.1089866767 |
- |
- |
hypothetical protein B456_007G3219002, partial [Gossypium raimondii] |
| 8 |
Hb_005373_030 |
0.1150317526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001529_010 |
0.1190104558 |
- |
- |
- |
| 10 |
Hb_016120_030 |
0.1198183244 |
- |
- |
hypothetical protein EUGRSUZ_D00080 [Eucalyptus grandis] |
| 11 |
Hb_033153_030 |
0.1200372437 |
- |
- |
PREDICTED: uncharacterized protein LOC105650145 [Jatropha curcas] |
| 12 |
Hb_003058_090 |
0.1283446003 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 13 |
Hb_007426_100 |
0.1316916179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_039946_040 |
0.1404335196 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 15 |
Hb_002874_100 |
0.1409262218 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000445_480 |
0.1469763664 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_004319_100 |
0.1474401708 |
- |
- |
PREDICTED: gibberellin-regulated protein 14 isoform X3 [Jatropha curcas] |
| 18 |
Hb_060236_010 |
0.1508733415 |
- |
- |
hypothetical protein JCGZ_26099 [Jatropha curcas] |
| 19 |
Hb_001091_010 |
0.1559224694 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X2 [Malus domestica] |
| 20 |
Hb_005245_030 |
0.1564413825 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g07570 [Jatropha curcas] |