| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
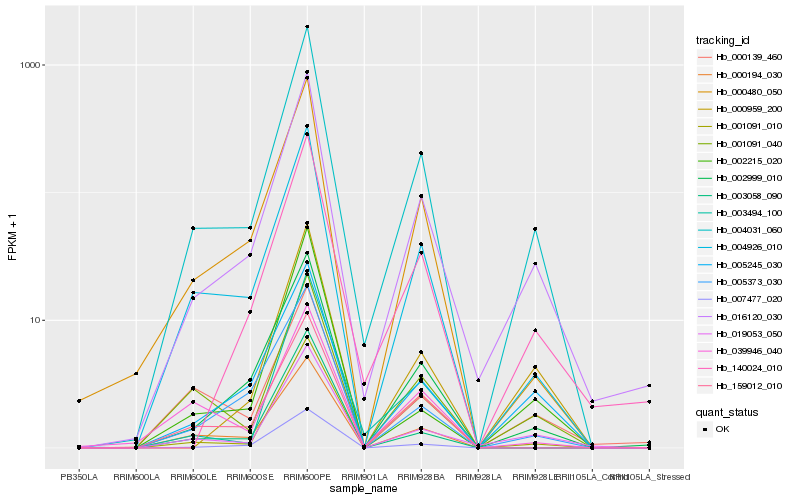
Hb_004031_060 |
0.0 |
- |
- |
hypothetical protein B456_007G3219002, partial [Gossypium raimondii] |
| 2 |
Hb_016120_030 |
0.0648236549 |
- |
- |
hypothetical protein EUGRSUZ_D00080 [Eucalyptus grandis] |
| 3 |
Hb_004926_010 |
0.0838483422 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 4 |
Hb_002999_010 |
0.1073059021 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 5 |
Hb_019053_050 |
0.1081978863 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_000480_050 |
0.1089866767 |
- |
- |
hypothetical protein PHAVU_004G154200g [Phaseolus vulgaris] |
| 7 |
Hb_001091_040 |
0.1120701619 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 8 |
Hb_005245_030 |
0.1170493393 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g07570 [Jatropha curcas] |
| 9 |
Hb_140024_010 |
0.1263815086 |
- |
- |
PREDICTED: uncharacterized protein C167.05 [Vitis vinifera] |
| 10 |
Hb_000959_200 |
0.1301071341 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 11 |
Hb_159012_010 |
0.1304557343 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 12 |
Hb_007477_020 |
0.1325317666 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA5 [Jatropha curcas] |
| 13 |
Hb_005373_030 |
0.1349521583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000194_030 |
0.1360941876 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 15 |
Hb_003058_090 |
0.1387940573 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 16 |
Hb_003494_100 |
0.1443085089 |
transcription factor |
TF Family: zf-HD |
Mini zinc finger 2 isoform 1 [Theobroma cacao] |
| 17 |
Hb_002215_020 |
0.1449082893 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 18 |
Hb_001091_010 |
0.1456835654 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X2 [Malus domestica] |
| 19 |
Hb_000139_460 |
0.1495171036 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 20 |
Hb_039946_040 |
0.1501218636 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |