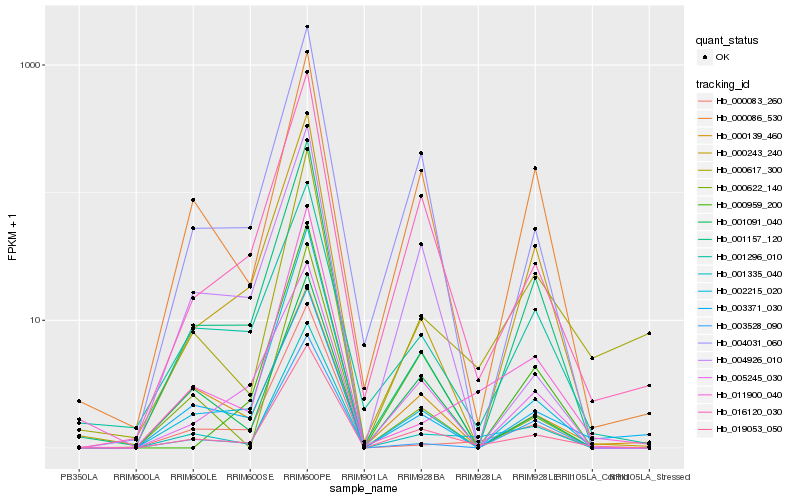
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019053_050 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_016120_030 |
0.1019165 |
- |
- |
hypothetical protein EUGRSUZ_D00080 [Eucalyptus grandis] |
| 3 |
Hb_001335_040 |
0.1076792222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004031_060 |
0.1081978863 |
- |
- |
hypothetical protein B456_007G3219002, partial [Gossypium raimondii] |
| 5 |
Hb_001091_040 |
0.1297592438 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 6 |
Hb_000086_530 |
0.1392529365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003371_030 |
0.1394601834 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005245_030 |
0.1421390928 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g07570 [Jatropha curcas] |
| 9 |
Hb_002215_020 |
0.1432579627 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 10 |
Hb_000959_200 |
0.1435822098 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 11 |
Hb_001157_120 |
0.1478233489 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 12 |
Hb_004926_010 |
0.1495103943 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 13 |
Hb_000139_460 |
0.1499102707 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 14 |
Hb_001296_010 |
0.1504836824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000243_240 |
0.1536239366 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 16 |
Hb_000622_140 |
0.156582099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000617_300 |
0.1584038743 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 18 |
Hb_003528_090 |
0.1634908827 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 19 |
Hb_011900_040 |
0.1653193581 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 20 |
Hb_000083_260 |
0.1656752265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |