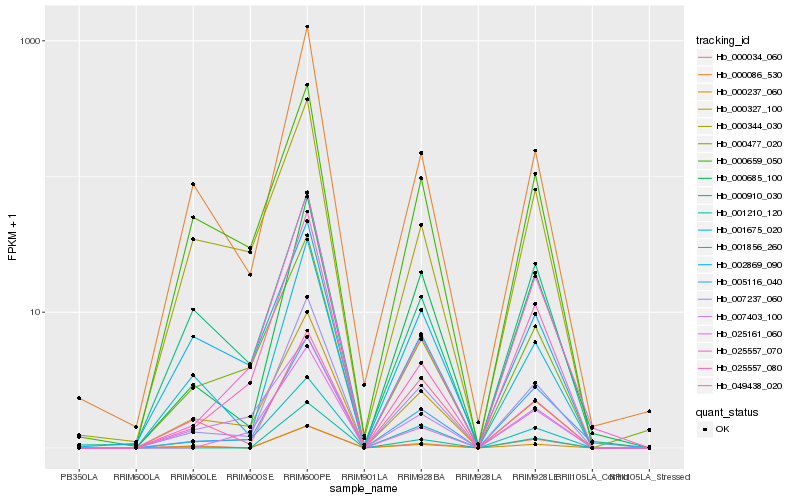
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007237_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_001856_260 |
0.088866475 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 3 |
Hb_001675_020 |
0.0962279503 |
- |
- |
- |
| 4 |
Hb_000327_100 |
0.1020595426 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 5 |
Hb_000086_530 |
0.1026344594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000237_060 |
0.1046365294 |
- |
- |
hypothetical protein PRUPE_ppa022052mg, partial [Prunus persica] |
| 7 |
Hb_000685_100 |
0.1091150561 |
- |
- |
hypothetical protein POPTR_0001s10630g [Populus trichocarpa] |
| 8 |
Hb_005116_040 |
0.1254412065 |
- |
- |
PREDICTED: uncharacterized protein LOC105131445 [Populus euphratica] |
| 9 |
Hb_000477_020 |
0.1272351151 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |
| 10 |
Hb_001210_120 |
0.12884636 |
- |
- |
hypothetical protein JCGZ_06226 [Jatropha curcas] |
| 11 |
Hb_025557_080 |
0.1309399284 |
transcription factor |
TF Family: zf-HD |
- |
| 12 |
Hb_000659_050 |
0.1340214649 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 13 |
Hb_025557_070 |
0.1345123173 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |
| 14 |
Hb_000344_030 |
0.1445570548 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 15 |
Hb_025161_060 |
0.1532646162 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 16 |
Hb_002869_090 |
0.1541382379 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 17 |
Hb_049438_020 |
0.1561801691 |
- |
- |
PREDICTED: peroxidase 20 [Jatropha curcas] |
| 18 |
Hb_000910_030 |
0.1567306081 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000034_060 |
0.1567555086 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 20 |
Hb_007403_100 |
0.1628275919 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |