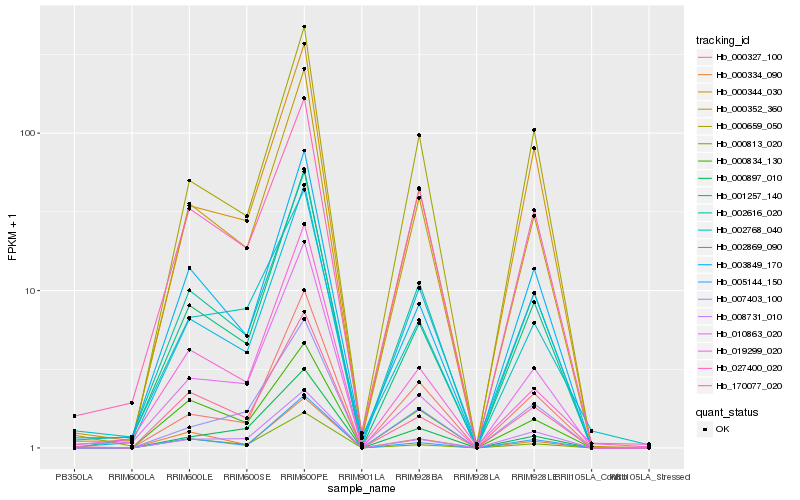
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_360 |
0.0 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 2 |
Hb_002616_020 |
0.0544035524 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 3 |
Hb_001257_140 |
0.0682143945 |
- |
- |
Gb:AAF02129.1, putative [Theobroma cacao] |
| 4 |
Hb_002869_090 |
0.0811008675 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 5 |
Hb_170077_020 |
0.0873184998 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 6 |
Hb_003849_170 |
0.0873613793 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 7 |
Hb_000327_100 |
0.0876773898 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 8 |
Hb_000334_090 |
0.1002249702 |
- |
- |
hypothetical protein JCGZ_16789 [Jatropha curcas] |
| 9 |
Hb_005144_150 |
0.1009600883 |
- |
- |
hypothetical protein VITISV_013572 [Vitis vinifera] |
| 10 |
Hb_000659_050 |
0.1016882996 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 11 |
Hb_000344_030 |
0.1044894236 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 12 |
Hb_019299_020 |
0.1055847954 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 13 |
Hb_000813_020 |
0.1074704556 |
- |
- |
PREDICTED: metacaspase-3-like [Jatropha curcas] |
| 14 |
Hb_027400_020 |
0.1089631992 |
- |
- |
PREDICTED: uncharacterized protein At5g22580 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000834_130 |
0.1117462664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008731_010 |
0.1119017659 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 17 |
Hb_010863_020 |
0.112381279 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000897_010 |
0.1128120643 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 19 |
Hb_002768_040 |
0.1157226798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007403_100 |
0.1158243625 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |