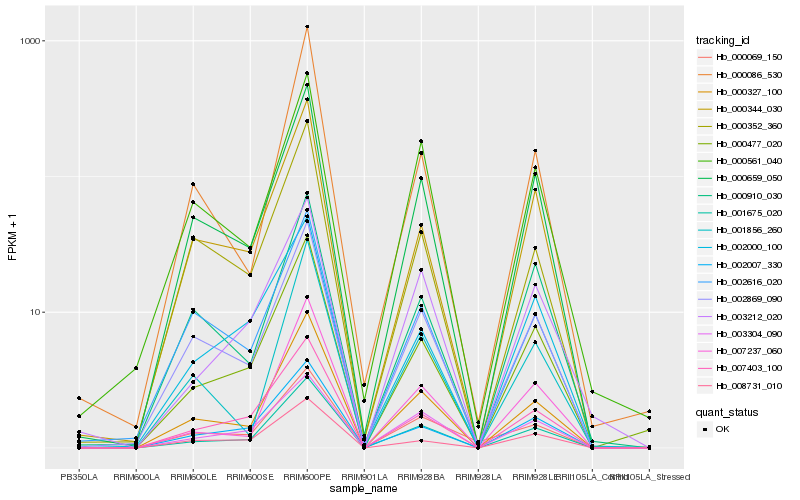
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001675_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000327_100 |
0.0550596503 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 3 |
Hb_000659_050 |
0.0754222253 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 4 |
Hb_000477_020 |
0.0765772611 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |
| 5 |
Hb_007403_100 |
0.0894055555 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 6 |
Hb_002869_090 |
0.0933773941 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 7 |
Hb_007237_060 |
0.0962279503 |
- |
- |
- |
| 8 |
Hb_002007_330 |
0.1001862371 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 9 |
Hb_000344_030 |
0.1016263277 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 10 |
Hb_003212_020 |
0.1038522705 |
- |
- |
hypothetical protein PHAVU_009G114800g [Phaseolus vulgaris] |
| 11 |
Hb_003304_090 |
0.104443778 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 12 |
Hb_000561_040 |
0.1171755521 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 13 |
Hb_002000_100 |
0.1242378895 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 14 |
Hb_001856_260 |
0.1243987485 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 15 |
Hb_000352_360 |
0.1278243237 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 16 |
Hb_008731_010 |
0.1278447773 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 17 |
Hb_000069_150 |
0.128466874 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 18 |
Hb_002616_020 |
0.1324834595 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 19 |
Hb_000910_030 |
0.1327013593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000086_530 |
0.1330756033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |