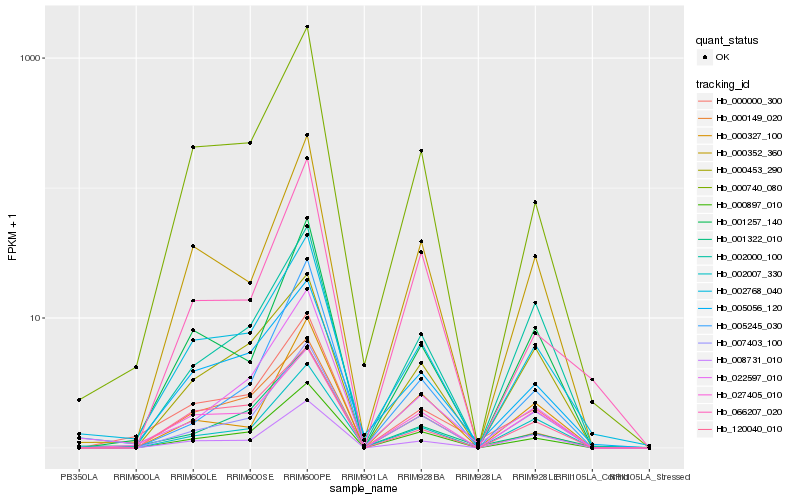
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000897_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 2 |
Hb_007403_100 |
0.0828690629 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 3 |
Hb_000740_080 |
0.0957290062 |
- |
- |
- |
| 4 |
Hb_002768_040 |
0.1095849288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000149_020 |
0.1096153666 |
- |
- |
Proline-rich receptor-like protein kinase PERK14 [Glycine soja] |
| 6 |
Hb_000352_360 |
0.1128120643 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 7 |
Hb_002007_330 |
0.1164990526 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 8 |
Hb_022597_010 |
0.1209221069 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 9 |
Hb_027405_010 |
0.1243541357 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 10 |
Hb_066207_020 |
0.1274777306 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 11 |
Hb_008731_010 |
0.1294299153 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 12 |
Hb_000327_100 |
0.1304531658 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 13 |
Hb_001322_010 |
0.1309290255 |
- |
- |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 14 |
Hb_000453_290 |
0.1319506055 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 15 |
Hb_000000_300 |
0.1319934058 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 16 |
Hb_005245_030 |
0.1360905531 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g07570 [Jatropha curcas] |
| 17 |
Hb_002000_100 |
0.1365155079 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 18 |
Hb_001257_140 |
0.1370658296 |
- |
- |
Gb:AAF02129.1, putative [Theobroma cacao] |
| 19 |
Hb_120040_010 |
0.1378907583 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 20 |
Hb_005056_120 |
0.1383145753 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |