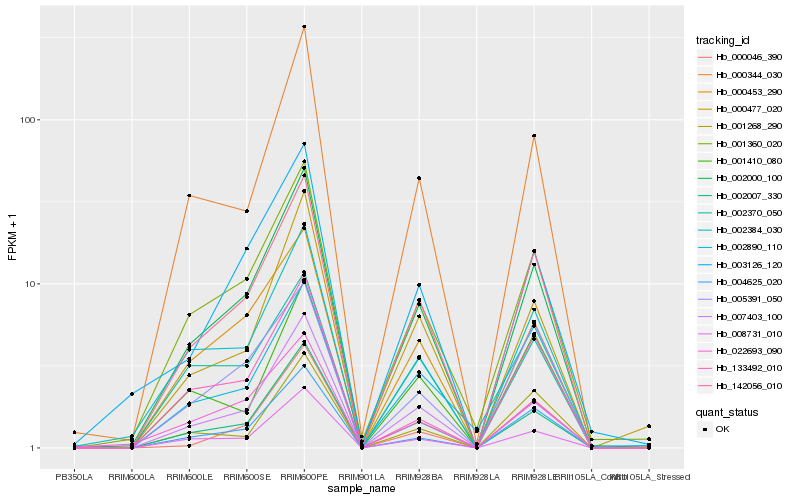
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142056_010 |
0.0 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002000_100 |
0.0562489187 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 3 |
Hb_001360_020 |
0.0860216797 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 4 |
Hb_002007_330 |
0.0881625377 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 5 |
Hb_005391_050 |
0.0978923369 |
- |
- |
PREDICTED: uncharacterized protein LOC105643654 [Jatropha curcas] |
| 6 |
Hb_000453_290 |
0.1005539034 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 7 |
Hb_002384_030 |
0.100901292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008731_010 |
0.1025615978 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 9 |
Hb_004625_020 |
0.1036457852 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 10 |
Hb_007403_100 |
0.1038383868 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 11 |
Hb_000344_030 |
0.1128271968 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 12 |
Hb_002370_050 |
0.1194556443 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 13 |
Hb_000477_020 |
0.1203233789 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |
| 14 |
Hb_022693_090 |
0.1235110259 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001410_080 |
0.1250114811 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_001268_290 |
0.1250854063 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 17 |
Hb_002890_110 |
0.125287938 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 18 |
Hb_003126_120 |
0.1271712013 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 19 |
Hb_000046_390 |
0.1277236691 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 20 |
Hb_133492_010 |
0.1283107132 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |