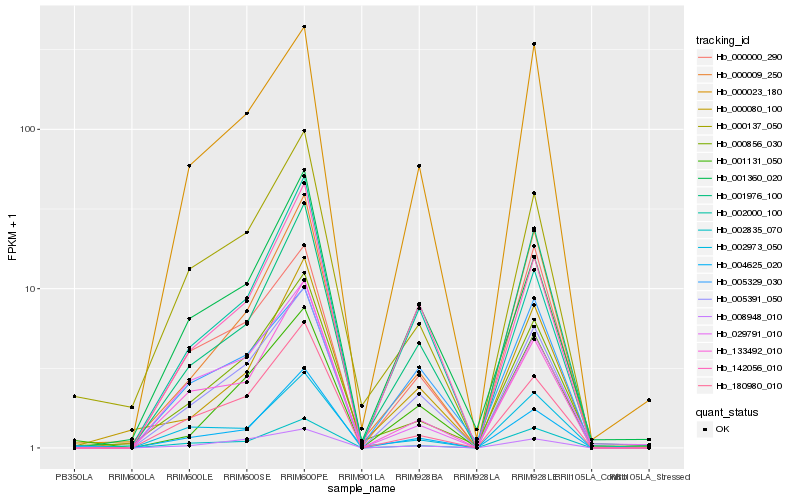
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005391_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643654 [Jatropha curcas] |
| 2 |
Hb_000023_180 |
0.0838419308 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 3 |
Hb_008948_010 |
0.0956113056 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 4 |
Hb_142056_010 |
0.0978923369 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002835_070 |
0.0992497707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001131_050 |
0.1098050982 |
- |
- |
hypothetical protein EUGRSUZ_C02968 [Eucalyptus grandis] |
| 7 |
Hb_001976_100 |
0.1104729194 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_180980_010 |
0.1142992257 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 9 |
Hb_133492_010 |
0.1168099565 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_004625_020 |
0.1168940917 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 11 |
Hb_000009_250 |
0.1198875218 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 12 |
Hb_005329_030 |
0.1205572339 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 13 |
Hb_000856_030 |
0.1231503437 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000137_050 |
0.1296460612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001360_020 |
0.1312212891 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 16 |
Hb_000080_100 |
0.1332065257 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 17 |
Hb_002973_050 |
0.1334417429 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 18 |
Hb_000000_290 |
0.1349727472 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 19 |
Hb_029791_010 |
0.1349738849 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 20 |
Hb_002000_100 |
0.1358805091 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |