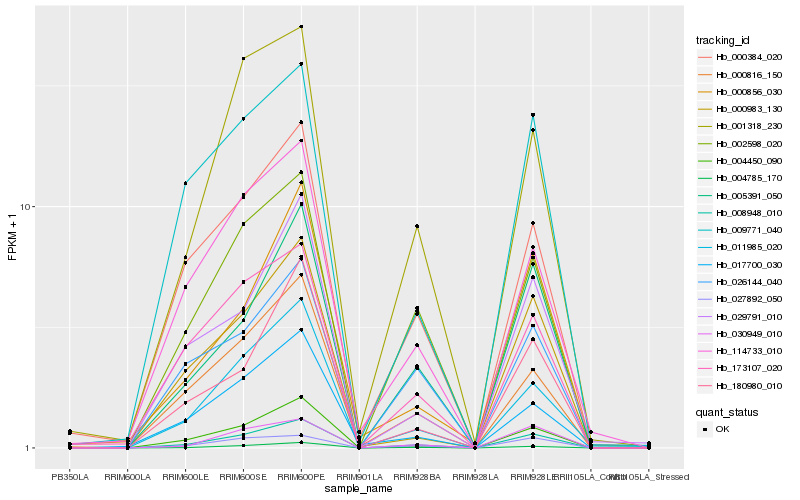
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008948_010 |
0.0 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004785_170 |
0.0880555437 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 3 |
Hb_005391_050 |
0.0956113056 |
- |
- |
PREDICTED: uncharacterized protein LOC105643654 [Jatropha curcas] |
| 4 |
Hb_002598_020 |
0.1081968435 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |
| 5 |
Hb_011985_020 |
0.1100822211 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004450_090 |
0.1112995647 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 7 |
Hb_000816_150 |
0.1113378411 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 8 |
Hb_030949_010 |
0.1164965094 |
- |
- |
hypothetical protein POPTR_0001s24140g [Populus trichocarpa] |
| 9 |
Hb_180980_010 |
0.1177768245 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 10 |
Hb_000384_020 |
0.1211586013 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |
| 11 |
Hb_000983_130 |
0.1216872149 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_027892_050 |
0.1219210831 |
- |
- |
hypothetical protein PHAVU_009G226400g [Phaseolus vulgaris] |
| 13 |
Hb_029791_010 |
0.1231602914 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 14 |
Hb_001318_230 |
0.1267523528 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 15 |
Hb_026144_040 |
0.1310683675 |
- |
- |
- |
| 16 |
Hb_009771_040 |
0.1325421954 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 17 |
Hb_173107_020 |
0.1332179435 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 18 |
Hb_114733_010 |
0.1334929578 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 19 |
Hb_017700_030 |
0.1339881782 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 20 |
Hb_000856_030 |
0.1354552299 |
- |
- |
ATP binding protein, putative [Ricinus communis] |