| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
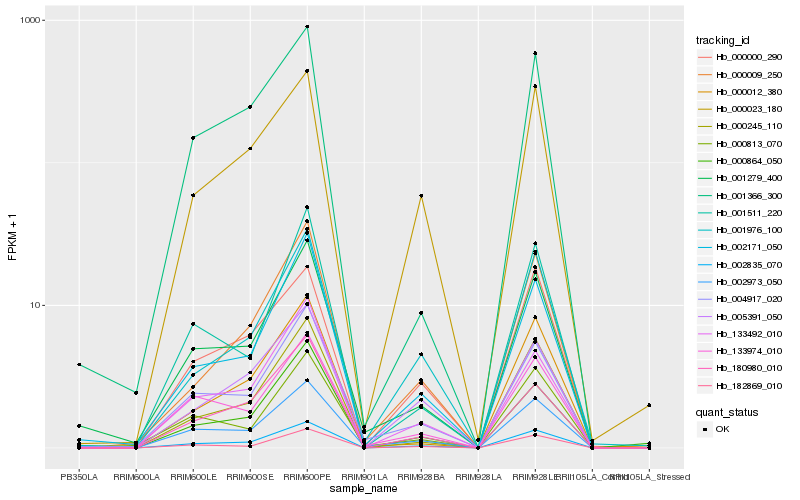
Hb_002835_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002973_050 |
0.0665929014 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 3 |
Hb_000023_180 |
0.0846375875 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 4 |
Hb_133974_010 |
0.092351202 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 5 |
Hb_004917_020 |
0.0930874058 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 6 |
Hb_182869_010 |
0.0938587478 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 7 |
Hb_000000_290 |
0.0944804739 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 8 |
Hb_001976_100 |
0.0948752503 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002171_050 |
0.0954413608 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 10 |
Hb_133492_010 |
0.0974004315 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_001279_400 |
0.0976900726 |
- |
- |
PREDICTED: uncharacterized protein LOC105633544 [Jatropha curcas] |
| 12 |
Hb_005391_050 |
0.0992497707 |
- |
- |
PREDICTED: uncharacterized protein LOC105643654 [Jatropha curcas] |
| 13 |
Hb_000009_250 |
0.1000367945 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 14 |
Hb_000245_110 |
0.1020113621 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 15 |
Hb_001366_300 |
0.1109831913 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 16 |
Hb_000864_050 |
0.1160839227 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_180980_010 |
0.1162828608 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 18 |
Hb_001511_220 |
0.1190334291 |
- |
- |
hypothetical protein POPTR_0008s06990g [Populus trichocarpa] |
| 19 |
Hb_000813_070 |
0.1195777887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000012_380 |
0.1199049007 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |