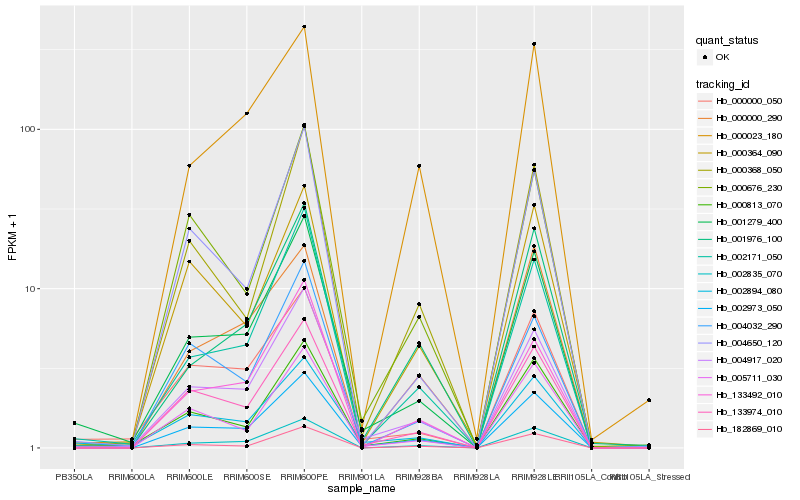
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002973_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 2 |
Hb_133974_010 |
0.0509013344 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 3 |
Hb_004917_020 |
0.0571751422 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 4 |
Hb_002835_070 |
0.0665929014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001279_400 |
0.0817295034 |
- |
- |
PREDICTED: uncharacterized protein LOC105633544 [Jatropha curcas] |
| 6 |
Hb_005711_030 |
0.0954246278 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 7 |
Hb_000813_070 |
0.0971927249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_182869_010 |
0.0999999773 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_000676_230 |
0.1067891508 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 10 |
Hb_002171_050 |
0.1090563907 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 11 |
Hb_000364_090 |
0.1094009086 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X2 [Jatropha curcas] |
| 12 |
Hb_133492_010 |
0.1115247314 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_000368_050 |
0.1126964419 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 14 |
Hb_000000_050 |
0.1127084384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002894_080 |
0.1141823266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000023_180 |
0.1146333128 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 17 |
Hb_004032_290 |
0.1156077727 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_004650_120 |
0.1157262723 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 19 |
Hb_000000_290 |
0.1181858883 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 20 |
Hb_001976_100 |
0.1185689713 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |