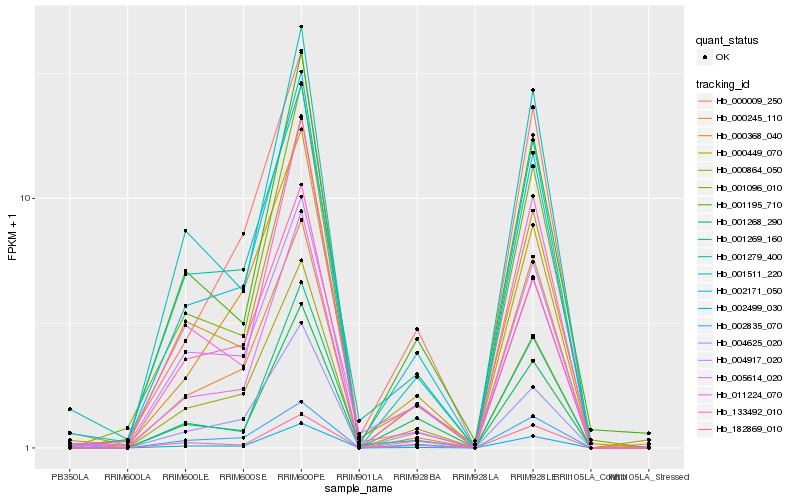
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002171_050 |
0.0 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 2 |
Hb_000449_070 |
0.078283566 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_004625_020 |
0.0812047432 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 4 |
Hb_004917_020 |
0.0830040588 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 5 |
Hb_005614_020 |
0.0833008822 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_133492_010 |
0.0836308653 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_002499_030 |
0.0841996587 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 8 |
Hb_001279_400 |
0.0861719317 |
- |
- |
PREDICTED: uncharacterized protein LOC105633544 [Jatropha curcas] |
| 9 |
Hb_001195_710 |
0.0902028646 |
- |
- |
glycosyltransferase family GT8 protein [Populus deltoides] |
| 10 |
Hb_001096_010 |
0.0907130689 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |
| 11 |
Hb_000009_250 |
0.0936687653 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 12 |
Hb_000368_040 |
0.0937925177 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000864_050 |
0.0951430569 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_001511_220 |
0.0951480703 |
- |
- |
hypothetical protein POPTR_0008s06990g [Populus trichocarpa] |
| 15 |
Hb_002835_070 |
0.0954413608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_182869_010 |
0.1005624715 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 17 |
Hb_011224_070 |
0.1033481943 |
- |
- |
PREDICTED: solute carrier family 35 member E3 [Jatropha curcas] |
| 18 |
Hb_000245_110 |
0.1058968629 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 19 |
Hb_001268_290 |
0.1079615315 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 20 |
Hb_001269_160 |
0.1085771659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |