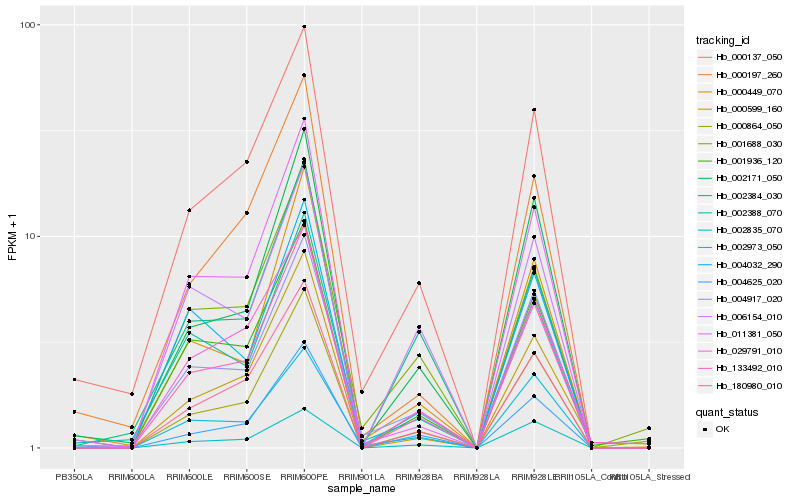
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_133492_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_180980_010 |
0.0533463536 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 3 |
Hb_000864_050 |
0.0783431229 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_004625_020 |
0.0816644398 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 5 |
Hb_002171_050 |
0.0836308653 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 6 |
Hb_004917_020 |
0.0936523823 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 7 |
Hb_029791_010 |
0.0937780774 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 8 |
Hb_011381_050 |
0.0958261818 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 9 |
Hb_002835_070 |
0.0974004315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000599_160 |
0.0982842199 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 11 |
Hb_000137_050 |
0.0997581229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001936_120 |
0.1020432823 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000449_070 |
0.1026787067 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_004032_290 |
0.1077279712 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_001688_030 |
0.108493957 |
- |
- |
hypothetical protein JCGZ_10716 [Jatropha curcas] |
| 16 |
Hb_002384_030 |
0.1103017449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006154_010 |
0.11094937 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_002388_070 |
0.1112550854 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_002973_050 |
0.1115247314 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 20 |
Hb_000197_260 |
0.1125628498 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |