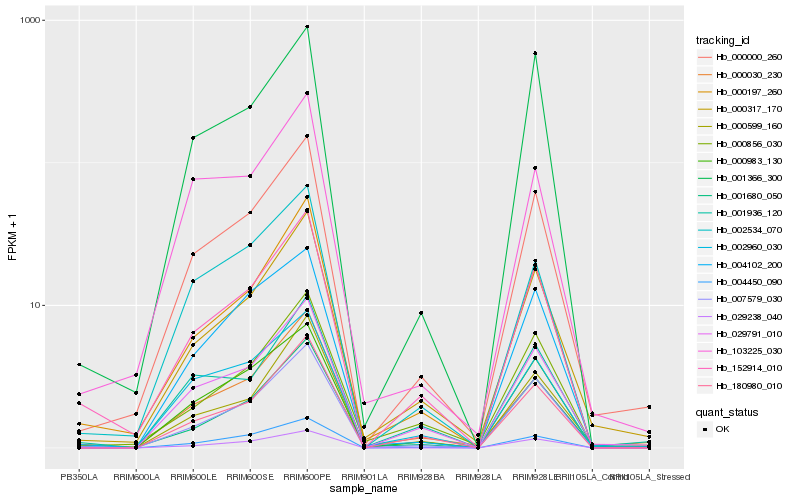
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_260 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 2 |
Hb_000317_170 |
0.0661389646 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 3 |
Hb_029791_010 |
0.0688231067 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 4 |
Hb_001680_050 |
0.0894634732 |
- |
- |
- |
| 5 |
Hb_007579_030 |
0.0926447435 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000983_130 |
0.0928909466 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_152914_010 |
0.0935812429 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 8 |
Hb_004450_090 |
0.0942267916 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 9 |
Hb_000197_260 |
0.0942875225 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 10 |
Hb_000030_230 |
0.095171058 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 11 |
Hb_000599_160 |
0.1006561602 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 12 |
Hb_180980_010 |
0.1011211452 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 13 |
Hb_004102_200 |
0.1043515976 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_002960_030 |
0.1047898987 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 15 |
Hb_000856_030 |
0.1052396494 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_002534_070 |
0.1061499189 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 17 |
Hb_103225_030 |
0.1065004333 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_001936_120 |
0.1073433103 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 19 |
Hb_029238_040 |
0.1078681774 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 20 |
Hb_001366_300 |
0.1089042622 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |