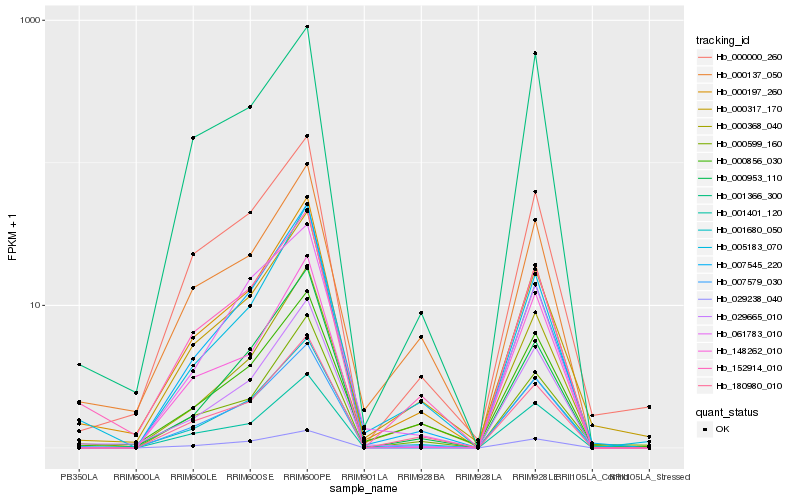
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001680_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_007579_030 |
0.062803063 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000317_170 |
0.066342082 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 4 |
Hb_029665_010 |
0.068768923 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000368_040 |
0.0692543218 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000197_260 |
0.0760037853 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 7 |
Hb_000856_030 |
0.0779092373 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_000000_260 |
0.0894634732 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 9 |
Hb_007545_220 |
0.0969904777 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 10 |
Hb_005183_070 |
0.0992993629 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 11 |
Hb_001401_120 |
0.100681834 |
- |
- |
hypothetical protein JCGZ_04273 [Jatropha curcas] |
| 12 |
Hb_000599_160 |
0.1012930685 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 13 |
Hb_180980_010 |
0.1026723872 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 14 |
Hb_152914_010 |
0.1048272019 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 15 |
Hb_000953_110 |
0.1063113105 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 16 |
Hb_029238_040 |
0.10861888 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 17 |
Hb_000137_050 |
0.1113924863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_061783_010 |
0.1121320709 |
- |
- |
PREDICTED: uncharacterized protein LOC105124983 isoform X4 [Populus euphratica] |
| 19 |
Hb_148262_010 |
0.112624191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001366_300 |
0.1133362786 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |