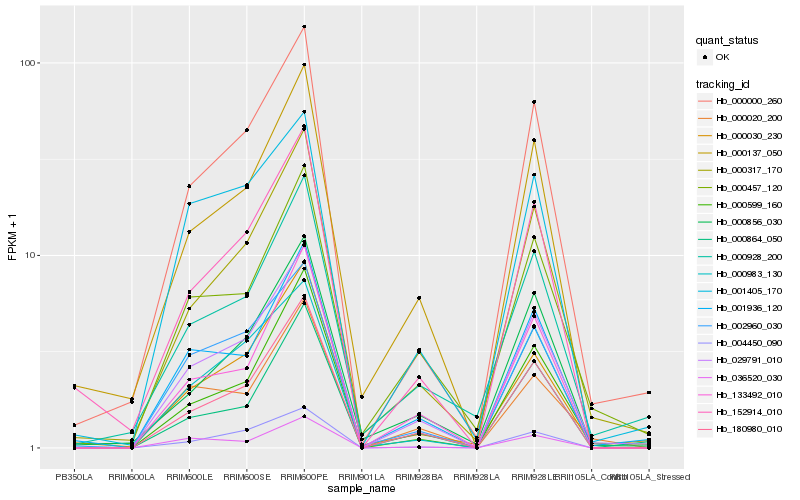
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029791_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 2 |
Hb_000000_260 |
0.0688231067 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 3 |
Hb_180980_010 |
0.0745546225 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 4 |
Hb_000317_170 |
0.080470574 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 5 |
Hb_001936_120 |
0.0840661764 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 6 |
Hb_133492_010 |
0.0937780774 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_000599_160 |
0.0949675485 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 8 |
Hb_002960_030 |
0.0989431957 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 9 |
Hb_004450_090 |
0.1014673575 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 10 |
Hb_000983_130 |
0.101523268 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_152914_010 |
0.1082102347 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 12 |
Hb_000457_120 |
0.1091053201 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 13 |
Hb_001405_170 |
0.110012789 |
- |
- |
aspartyl protease family protein [Populus trichocarpa] |
| 14 |
Hb_000137_050 |
0.1108557587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000020_200 |
0.1116350502 |
- |
- |
hypothetical protein EUGRSUZ_B00411 [Eucalyptus grandis] |
| 16 |
Hb_000030_230 |
0.111679715 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 17 |
Hb_000864_050 |
0.1123404317 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000856_030 |
0.1135752318 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_000928_200 |
0.1136215928 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 20 |
Hb_036520_030 |
0.1139732059 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |