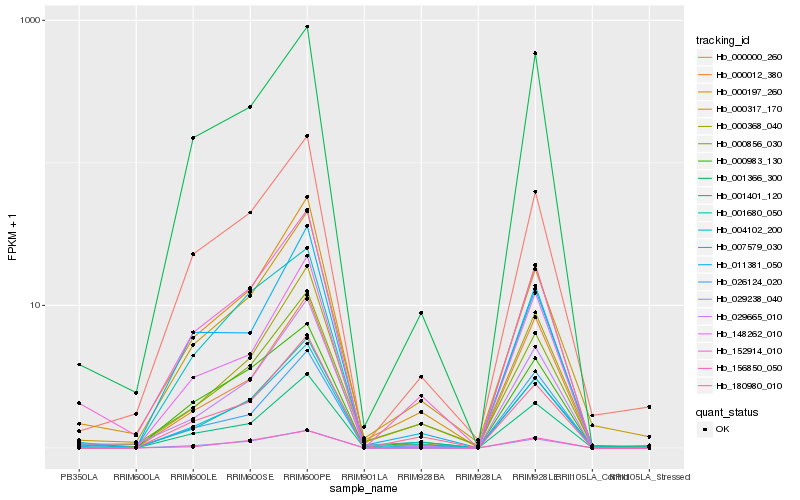
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007579_030 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_001680_050 |
0.062803063 |
- |
- |
- |
| 3 |
Hb_001401_120 |
0.0738108038 |
- |
- |
hypothetical protein JCGZ_04273 [Jatropha curcas] |
| 4 |
Hb_029665_010 |
0.0773991488 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001366_300 |
0.0786034717 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 6 |
Hb_029238_040 |
0.0791640903 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 7 |
Hb_000197_260 |
0.0831315689 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 8 |
Hb_148262_010 |
0.0897886884 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000000_260 |
0.0926447435 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 10 |
Hb_152914_010 |
0.0928461101 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 11 |
Hb_000368_040 |
0.0933375382 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000983_130 |
0.0940362387 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000317_170 |
0.096654078 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 14 |
Hb_180980_010 |
0.0994294648 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 15 |
Hb_000856_030 |
0.1014668373 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_026124_020 |
0.101931874 |
- |
- |
PREDICTED: uncharacterized protein LOC105644241 isoform X1 [Jatropha curcas] |
| 17 |
Hb_156850_050 |
0.1029470072 |
- |
- |
PREDICTED: uncharacterized protein LOC105635250 [Jatropha curcas] |
| 18 |
Hb_000012_380 |
0.1035086225 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011381_050 |
0.1039986809 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 20 |
Hb_004102_200 |
0.1044230175 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |