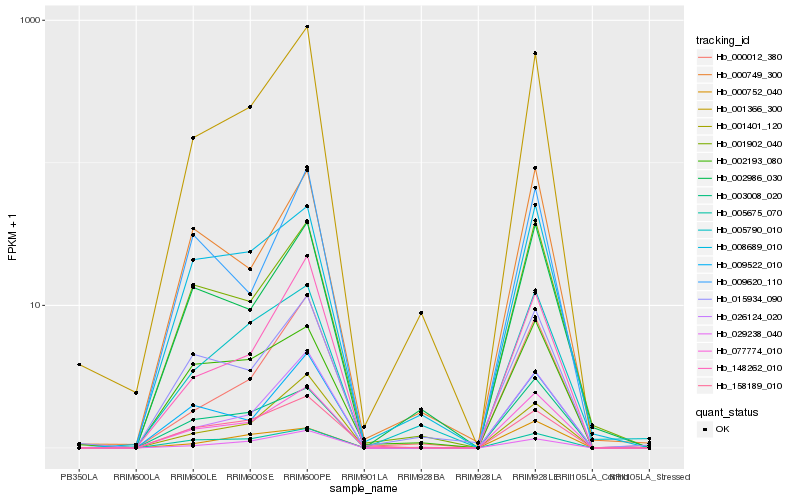
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077774_010 |
0.0 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 2 |
Hb_001366_300 |
0.0773505973 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 3 |
Hb_003008_020 |
0.0898913659 |
- |
- |
auxin-responsive family protein [Populus trichocarpa] |
| 4 |
Hb_026124_020 |
0.0932657152 |
- |
- |
PREDICTED: uncharacterized protein LOC105644241 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001902_040 |
0.0984843366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005675_070 |
0.1039777392 |
- |
- |
hypothetical protein EUGRSUZ_K01585 [Eucalyptus grandis] |
| 7 |
Hb_009522_010 |
0.1043600359 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 8 |
Hb_008689_010 |
0.1133908183 |
- |
- |
hypothetical protein POPTR_0006s25330g [Populus trichocarpa] |
| 9 |
Hb_002986_030 |
0.1136836381 |
- |
- |
hypothetical protein JCGZ_12380 [Jatropha curcas] |
| 10 |
Hb_148262_010 |
0.1175067191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001401_120 |
0.1194285334 |
- |
- |
hypothetical protein JCGZ_04273 [Jatropha curcas] |
| 12 |
Hb_015934_090 |
0.120729545 |
- |
- |
PREDICTED: uncharacterized protein LOC105640158 [Jatropha curcas] |
| 13 |
Hb_000749_300 |
0.1225226638 |
- |
- |
PREDICTED: uncharacterized protein LOC105129180 [Populus euphratica] |
| 14 |
Hb_000012_380 |
0.1226499479 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_158189_010 |
0.1254643617 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 16 |
Hb_029238_040 |
0.1260170126 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 17 |
Hb_000752_040 |
0.1317443438 |
- |
- |
PREDICTED: uncharacterized protein LOC104884990 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_005790_010 |
0.1326682757 |
- |
- |
hypothetical protein POPTR_0004s24080g [Populus trichocarpa] |
| 19 |
Hb_002193_080 |
0.1340176919 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_009620_110 |
0.134228053 |
- |
- |
- |