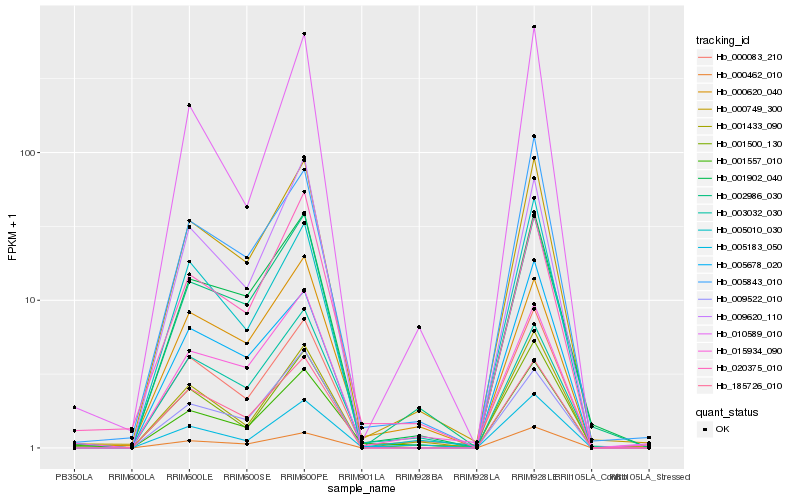
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000749_300 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105129180 [Populus euphratica] |
| 2 |
Hb_001902_040 |
0.0582527924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002986_030 |
0.0632863486 |
- |
- |
hypothetical protein JCGZ_12380 [Jatropha curcas] |
| 4 |
Hb_000083_210 |
0.0690317975 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 5 |
Hb_185726_010 |
0.0728071947 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 6 |
Hb_000462_010 |
0.0818452057 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |
| 7 |
Hb_015934_090 |
0.0868365392 |
- |
- |
PREDICTED: uncharacterized protein LOC105640158 [Jatropha curcas] |
| 8 |
Hb_001500_130 |
0.0868864441 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 9 |
Hb_005010_030 |
0.0884421306 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 10 |
Hb_003032_030 |
0.0930189433 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009620_110 |
0.0939818149 |
- |
- |
- |
| 12 |
Hb_001433_090 |
0.094688447 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 13 |
Hb_005843_010 |
0.0955473374 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 14 |
Hb_005183_050 |
0.1003857767 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 15 |
Hb_001557_010 |
0.101425936 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 16 |
Hb_000620_040 |
0.1017259236 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_009522_010 |
0.1021767321 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 18 |
Hb_005678_020 |
0.1027485222 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_020375_010 |
0.1029584812 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 20 |
Hb_010589_010 |
0.1039026242 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |