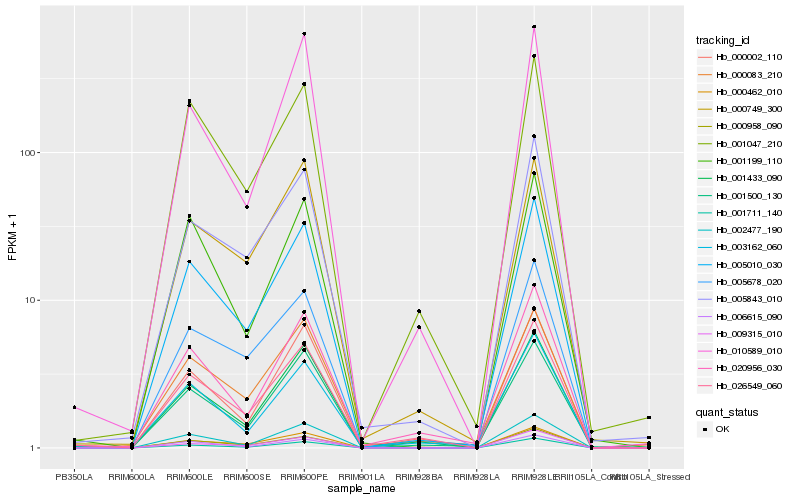
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005010_030 |
0.0 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 2 |
Hb_001711_140 |
0.0553794919 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 3 |
Hb_002477_190 |
0.0571246002 |
- |
- |
hypothetical protein EUGRSUZ_C03380, partial [Eucalyptus grandis] |
| 4 |
Hb_000462_010 |
0.0586992182 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |
| 5 |
Hb_006615_090 |
0.0679187978 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 6 |
Hb_005678_020 |
0.0689461118 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_005843_010 |
0.0718487962 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 8 |
Hb_001047_210 |
0.0739979632 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 9 |
Hb_001433_090 |
0.0740471347 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 10 |
Hb_001500_130 |
0.0789154796 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 11 |
Hb_026549_060 |
0.0793248577 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 12 |
Hb_009315_010 |
0.0806791465 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 13 |
Hb_000958_090 |
0.0835998881 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 14 |
Hb_001199_110 |
0.0848036938 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000083_210 |
0.0852638313 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 16 |
Hb_000749_300 |
0.0884421306 |
- |
- |
PREDICTED: uncharacterized protein LOC105129180 [Populus euphratica] |
| 17 |
Hb_000002_110 |
0.0920466487 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 18 |
Hb_020956_030 |
0.0932930169 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 19 |
Hb_003162_060 |
0.0941784214 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 20 |
Hb_010589_010 |
0.0962921313 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |