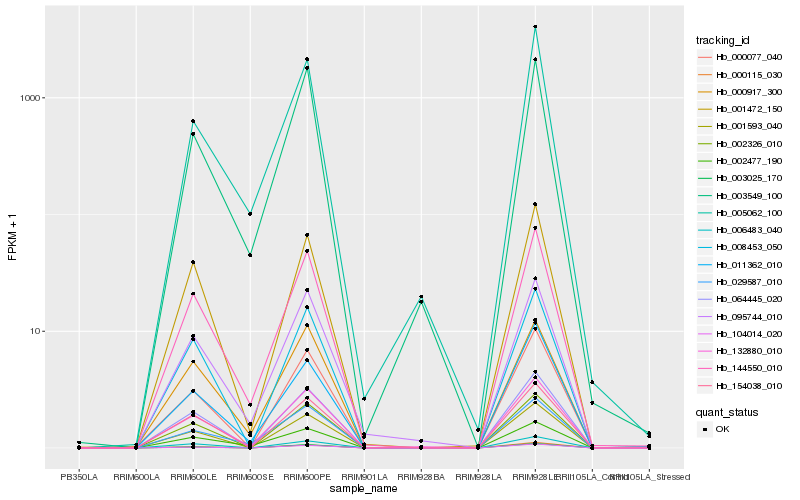
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_144550_010 |
0.0 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 2 |
Hb_132880_010 |
0.0630518323 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 3 |
Hb_001472_150 |
0.0674482177 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 4 |
Hb_029587_010 |
0.0682821937 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase [Glycine soja] |
| 5 |
Hb_001593_040 |
0.0723717513 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 6 |
Hb_095744_010 |
0.0725322679 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 7 |
Hb_064445_020 |
0.0739396285 |
- |
- |
PREDICTED: serine carboxypeptidase-like 34 [Populus euphratica] |
| 8 |
Hb_002477_190 |
0.0781804032 |
- |
- |
hypothetical protein EUGRSUZ_C03380, partial [Eucalyptus grandis] |
| 9 |
Hb_000077_040 |
0.080697237 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 10 |
Hb_008453_050 |
0.0807421795 |
- |
- |
PREDICTED: peroxidase 64 [Jatropha curcas] |
| 11 |
Hb_011362_010 |
0.0808670497 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 12 |
Hb_002326_010 |
0.0809914965 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 13 |
Hb_003549_100 |
0.0821538793 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 14 |
Hb_104014_020 |
0.0823302642 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000115_030 |
0.0826460097 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 42 [Jatropha curcas] |
| 16 |
Hb_005062_100 |
0.0835362584 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000917_300 |
0.0839820799 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 18 |
Hb_154038_010 |
0.0844948425 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_006483_040 |
0.0846036285 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 20 |
Hb_003025_170 |
0.08478597 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59900 [Jatropha curcas] |