| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
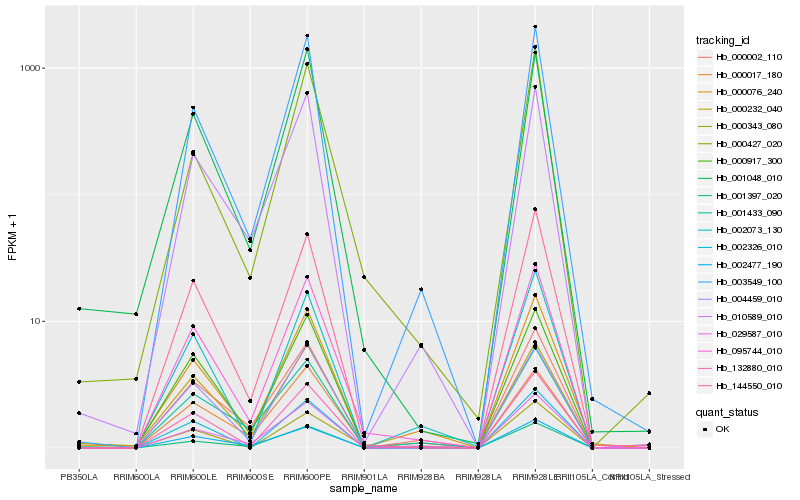
Hb_095744_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 2 |
Hb_132880_010 |
0.0619418545 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 3 |
Hb_003549_100 |
0.0688061866 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 4 |
Hb_004459_010 |
0.0719262588 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 5 |
Hb_144550_010 |
0.0725322679 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_000917_300 |
0.0751137429 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 7 |
Hb_010589_010 |
0.0788062903 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 8 |
Hb_029587_010 |
0.0789270924 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase [Glycine soja] |
| 9 |
Hb_000076_240 |
0.0840730351 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 10 |
Hb_001397_020 |
0.085483773 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 11 |
Hb_000343_080 |
0.092215947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002073_130 |
0.0924259619 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 13 |
Hb_000002_110 |
0.0941556095 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 14 |
Hb_001048_010 |
0.0943989089 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 7 [Jatropha curcas] |
| 15 |
Hb_002477_190 |
0.0947875937 |
- |
- |
hypothetical protein EUGRSUZ_C03380, partial [Eucalyptus grandis] |
| 16 |
Hb_000017_180 |
0.0957502751 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 17 |
Hb_000427_020 |
0.0967989852 |
- |
- |
PsbA, partial [Dianthus pygmaeus] |
| 18 |
Hb_001433_090 |
0.0982905945 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 19 |
Hb_000232_040 |
0.0987458289 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X2 [Populus euphratica] |
| 20 |
Hb_002326_010 |
0.098808838 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |