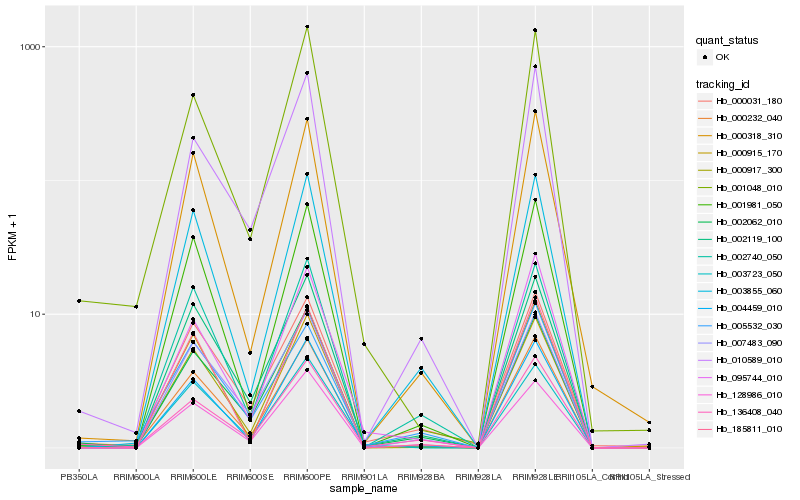
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000232_040 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X2 [Populus euphratica] |
| 2 |
Hb_000917_300 |
0.069359139 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 3 |
Hb_002062_010 |
0.0734881863 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 4 |
Hb_001048_010 |
0.0756058211 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 7 [Jatropha curcas] |
| 5 |
Hb_002119_100 |
0.0759995781 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 6 |
Hb_004459_010 |
0.0770709976 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 7 |
Hb_001981_050 |
0.0860177551 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 8 |
Hb_136408_040 |
0.0866265102 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 9 |
Hb_000318_310 |
0.0894161888 |
- |
- |
hypothetical protein POPTR_0009s11190g [Populus trichocarpa] |
| 10 |
Hb_000915_170 |
0.0899241432 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 11 |
Hb_000031_180 |
0.0925433349 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 12 |
Hb_005532_030 |
0.0949800722 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_007483_090 |
0.0957118018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003855_060 |
0.0958804191 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 15 |
Hb_185811_010 |
0.0964719396 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 16 |
Hb_128986_010 |
0.0970106959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003723_050 |
0.0972673645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010589_010 |
0.0973971323 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 19 |
Hb_095744_010 |
0.0987458289 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 20 |
Hb_002740_050 |
0.0990762744 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86-like [Populus euphratica] |