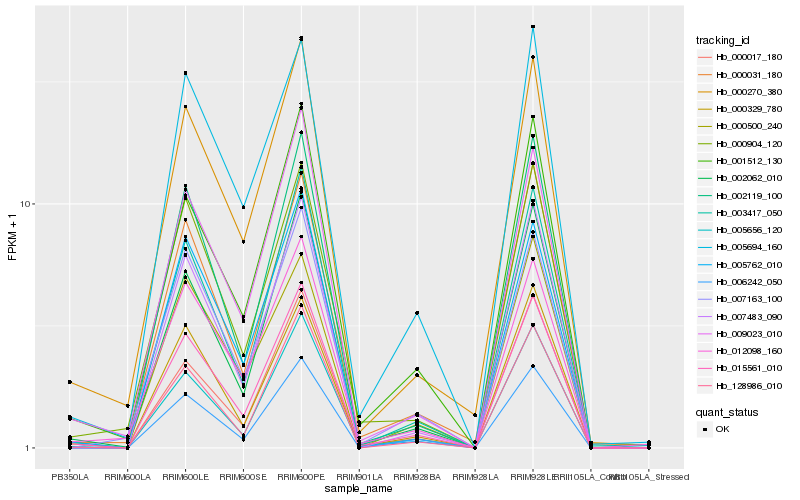
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015561_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002062_010 |
0.052429784 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 3 |
Hb_001512_130 |
0.0575254072 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 4 |
Hb_002119_100 |
0.064416929 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 5 |
Hb_009023_010 |
0.0651581872 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 6 |
Hb_003417_050 |
0.0680087322 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_007483_090 |
0.0702202548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005656_120 |
0.0735695805 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 9 |
Hb_006242_050 |
0.0746774896 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 10 |
Hb_000270_380 |
0.0749061995 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 11 |
Hb_128986_010 |
0.0751653673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000031_180 |
0.0762608989 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 13 |
Hb_000329_780 |
0.0768914245 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 14 |
Hb_000904_120 |
0.0809112767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000017_180 |
0.0816632946 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 16 |
Hb_012098_160 |
0.082637184 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 17 |
Hb_005762_010 |
0.0830484477 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_005694_160 |
0.0834703844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007163_100 |
0.0841032929 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 20 |
Hb_000500_240 |
0.0894146138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |