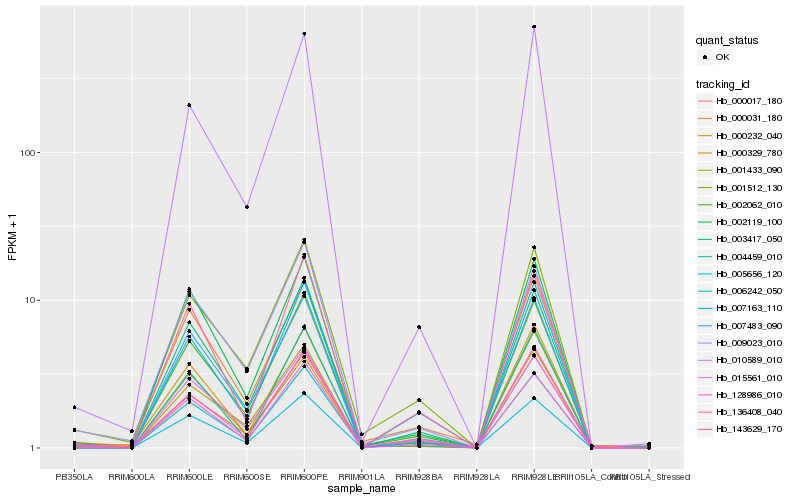
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002062_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 2 |
Hb_128986_010 |
0.0479959498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_015561_010 |
0.052429784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000017_180 |
0.0561443409 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 5 |
Hb_005656_120 |
0.0570344434 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 6 |
Hb_003417_050 |
0.0590476597 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002119_100 |
0.0595353143 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 8 |
Hb_001512_130 |
0.0610803959 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 9 |
Hb_007483_090 |
0.0637620401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_009023_010 |
0.0734044666 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 11 |
Hb_000232_040 |
0.0734881863 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X2 [Populus euphratica] |
| 12 |
Hb_136408_040 |
0.0737642939 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 13 |
Hb_007163_110 |
0.0776702665 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 14 |
Hb_004459_010 |
0.0812294243 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 15 |
Hb_006242_050 |
0.0812808741 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 16 |
Hb_000031_180 |
0.0815221651 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 17 |
Hb_143629_170 |
0.0819797135 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 18 |
Hb_010589_010 |
0.0862591862 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 19 |
Hb_001433_090 |
0.0864665527 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 20 |
Hb_000329_780 |
0.0875312168 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |