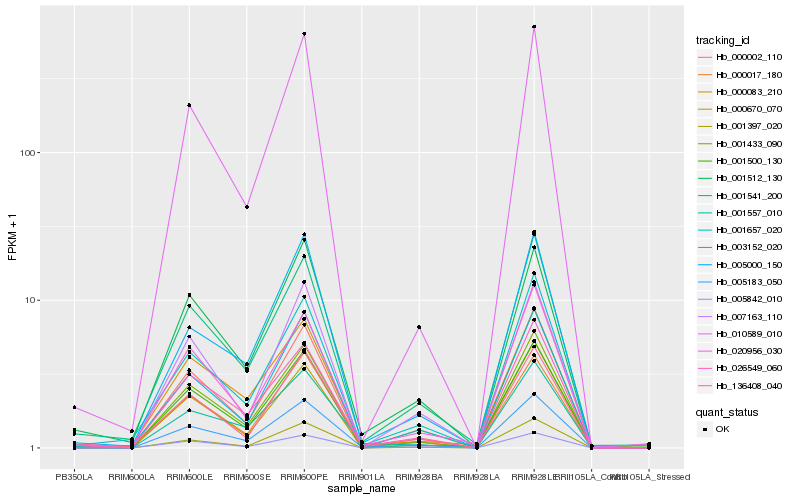
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005183_050 |
0.0 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 2 |
Hb_001433_090 |
0.0589079284 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 3 |
Hb_000017_180 |
0.0592977141 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 4 |
Hb_000002_110 |
0.0750046644 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 5 |
Hb_003152_020 |
0.0756328328 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 6 |
Hb_026549_060 |
0.0769686197 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 7 |
Hb_005842_010 |
0.0777089005 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 8 |
Hb_001657_020 |
0.078260248 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 9 |
Hb_001541_200 |
0.0803680618 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 10 |
Hb_001397_020 |
0.0807092489 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 11 |
Hb_010589_010 |
0.0812179466 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 12 |
Hb_005000_150 |
0.0823245061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001557_010 |
0.0859726576 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 14 |
Hb_001512_130 |
0.0885282831 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 15 |
Hb_007163_110 |
0.0888365028 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 16 |
Hb_136408_040 |
0.0892930379 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 17 |
Hb_020956_030 |
0.0910539551 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 18 |
Hb_001500_130 |
0.0911363728 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 19 |
Hb_000670_070 |
0.096578645 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 20 |
Hb_000083_210 |
0.0969166429 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |