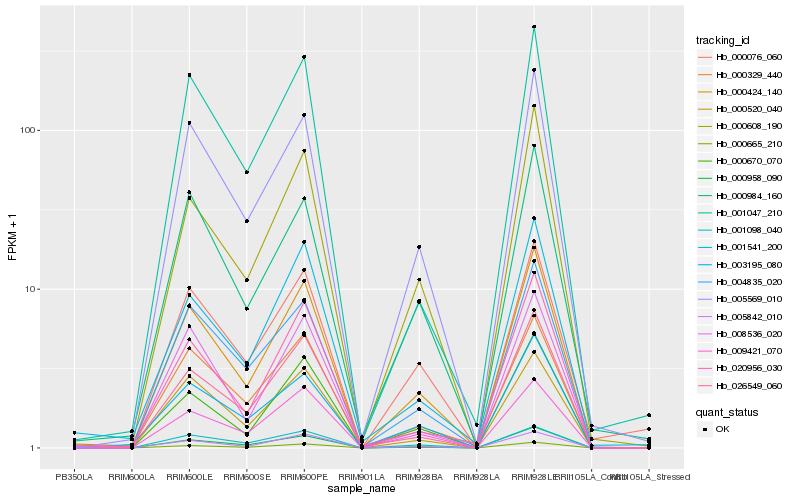
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000424_140 |
0.0 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_005569_010 |
0.0558732895 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 3 |
Hb_000670_070 |
0.0642395669 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 4 |
Hb_005842_010 |
0.0658843536 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 5 |
Hb_000958_090 |
0.0666672942 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 6 |
Hb_000608_190 |
0.0685587873 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 7 |
Hb_009421_070 |
0.0803718751 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 8 |
Hb_004835_020 |
0.0812863805 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 9 |
Hb_000329_440 |
0.0853256251 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 10 |
Hb_000520_040 |
0.0866078355 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 11 |
Hb_026549_060 |
0.0870097556 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 12 |
Hb_003195_080 |
0.0883977227 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 13 |
Hb_008536_020 |
0.089261479 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 14 |
Hb_000984_160 |
0.0926072145 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000665_210 |
0.0930100211 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 16 |
Hb_000076_060 |
0.093686521 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 17 |
Hb_001098_040 |
0.0947709051 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 18 |
Hb_001047_210 |
0.0966551286 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 19 |
Hb_020956_030 |
0.0983092528 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 20 |
Hb_001541_200 |
0.1025013249 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |