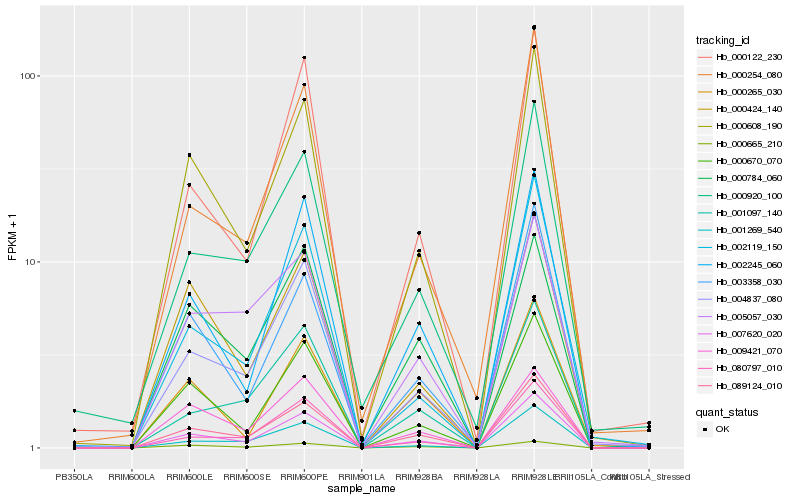
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089124_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 2 |
Hb_007620_020 |
0.039414038 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000608_190 |
0.072353154 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 4 |
Hb_004837_080 |
0.0845662099 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 5 |
Hb_000670_070 |
0.1007796412 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 6 |
Hb_080797_010 |
0.1020025841 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 7 |
Hb_003358_030 |
0.1020276688 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 8 |
Hb_001269_540 |
0.1027588511 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 9 |
Hb_002245_060 |
0.1040387721 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001097_140 |
0.1046646708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000254_080 |
0.1078585634 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 12 |
Hb_000920_100 |
0.1082156226 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000665_210 |
0.1110192981 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 14 |
Hb_000122_230 |
0.1114157645 |
- |
- |
laccase, putative [Ricinus communis] |
| 15 |
Hb_005057_030 |
0.1182306024 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 16 |
Hb_000424_140 |
0.1206353062 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_002119_150 |
0.1211361496 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 18 |
Hb_000265_030 |
0.1244382028 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 19 |
Hb_009421_070 |
0.125696805 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 20 |
Hb_000784_060 |
0.1278343586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |