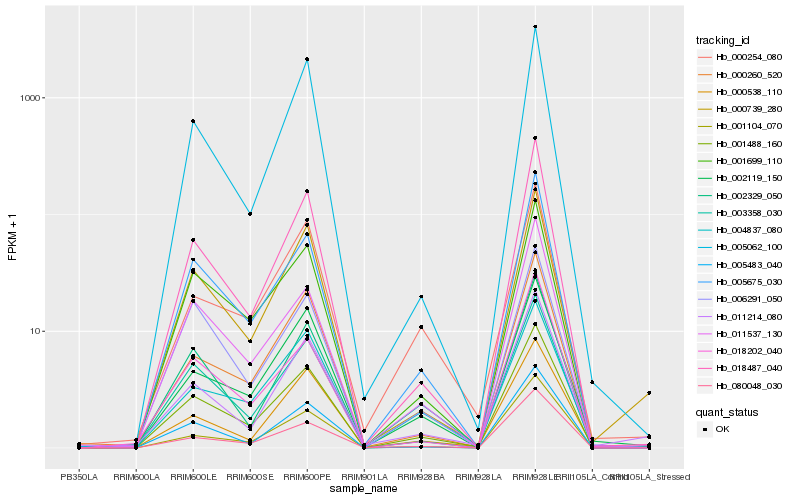
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_160 |
0.0 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_005675_030 |
0.0615118069 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 3 |
Hb_001699_110 |
0.0698198101 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 4 |
Hb_018487_040 |
0.0765421392 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 5 |
Hb_011537_130 |
0.0824568884 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_080048_030 |
0.0842539421 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 7 |
Hb_000739_280 |
0.0865908263 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_003358_030 |
0.0876235003 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 9 |
Hb_000260_520 |
0.0902683648 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_005483_040 |
0.0903928386 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 11 |
Hb_002119_150 |
0.0912923365 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 12 |
Hb_001104_070 |
0.0924749104 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 13 |
Hb_002329_050 |
0.0947669474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011214_080 |
0.0997604498 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 15 |
Hb_000254_080 |
0.1007985979 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 16 |
Hb_000538_110 |
0.1016364546 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 17 |
Hb_004837_080 |
0.1020916283 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 18 |
Hb_006291_050 |
0.1023194381 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_005062_100 |
0.1032640414 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_018202_040 |
0.1033577204 |
- |
- |
PREDICTED: phosphoenolpyruvate/phosphate translocator 2, chloroplastic-like [Jatropha curcas] |